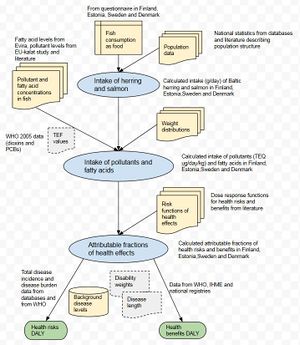
+ Show code- Hide code # This is code Op_en7748/hia on page [[Benefit-risk assessment of Baltic herring and salmon intake]]
library(OpasnetUtils)
library(ggplot2)
showLoctable <- function(name = ".GlobalEnv") {
loctable <- data.frame()
for(i in ls(name = name)) {
if(class(get(i)) == "ovariable") {
for(j in colnames(get(i)@output)) {
if(!(grepl("Source", j) | grepl("Result", j))) {
loctable <- rbind(
loctable,
data.frame(
Ovariable = i,
Index = j,
Class = class(get(i)@output[[j]]),
NumLoc = length(unique(get(i)@output[[j]])),
Locations = paste(head(unique(get(i)@output[[j]])), collapse = " ")
)
)
}
}
}
}
return(loctable)
}
objects.latest("Op_en7748", code_name = "bayes") #: pcd.pred, ans.pred, mu.pred
objects.latest("Op_en1838", code_name = "bayes") #nutrient concentrations
objects.latest("Op_en7748", code_name = "exposure")
# [[Benefit-risk assessment of Baltic herring and salmon intake]]
# ovariables expodiox, expoomegaherring
# expoomegaherring is EMPTY! Fix this.
# This should use the newer expodiox$Compound<- syntax.
expodiox@output$Exposure_agent <- "TEQ"
expodiox@marginal <- c(expodiox@marginal, TRUE)
exposure <- expodiox
# Run this when expoomegaherring works
#exposure <- combine(expodiox, expoomegaherring)
solet <- opbase.data("Op_fi3831.saantioletukset")
#!!++++++++++++++++++++++++++++++++++++++++++++++++++++++++++
## Background-exposure to vitamin D and omega-3
addexposure <- EvalOutput(Ovariable("addexposure", ddata = "Op_fi3831.tausta_altistus"))
#ii++++++++++++++++++++++++++++++++++++++++++++++++++++++++++
# Empty values ("") in indices must be replaced by NA so that Ops works correctly.
levels(addexposure@output$Sukupuoli)[levels(addexposure@output$Sukupuoli) == ""] <- NA
levels(addexposure@output$Exposure_agent)[levels(addexposure@output$Exposure_agent) == ""] <- NA
addexposure@output <- fillna(addexposure@output, c("Sukupuoli", "Exposure_agent"))
sumitem <- function(
ova, #ovariable that has locations to sum
cond, # index column that contains the locations to sum
condvalue, # vector of locations to sum
sumvalue # location to be given to the rows with the sums
) {
d <- ova
d@output <- d@output[d@output[[cond]] %in% condvalue , ]
d <- oapply(d, cols = cond, FUN = sum)
d@output[[cond]] <- sumvalue
ova@output <- orbind(ova, d)
return(ova)
}
addexposure <- sumitem(addexposure, "Exposure_agent", c("PCDDF", "PCB"), "TEQ")
addexposure <- sumitem(addexposure, "Exposure_agent", c("EPA", "DHA"), "Omega3")
addexposure <- unkeep(addexposure, prevresults = TRUE, sources = TRUE)
# Make the background exposure uncertain rather than an index.
#################### USE collapseMarginal instead. Produces ovariable expo.add
background <- Ovariable(
output = data.frame(
Iter = 1:get("N", envir = openv),
Tausta = c("Kyllä", "Ei")[sample(2, get("N", envir = openv), replace = TRUE)],
Result = 1
),
marginal = c(TRUE, FALSE, FALSE)
)
addexposure <- addexposure * background
# This should be done in data already? Background compatibility? #### YES DO IN DATA
colnames(addexposure@output)[1:2] <- c("Background", "Gender")
levels(addexposure@output$Gender) <- c("Male", "Female")
levels(addexposure@output$Background) <- c("No", "Yes")
exposure <- exposure + addexposure
################## CREATE a separate ovarible about children's exposure: expo.child
# Convert mother's dioxin intake (pg/d) into child's dioxin concentration (pg/g) in fat after 6 months of breast-feeding.
# For details, see [[ERF of dioxin#Dental defects]]
t0.5 <- Ovariable("t0.5", data = solet[solet$Muuttuja == "TEQ-puoliintumisaika" , ]["Result"])
f_ing <- Ovariable("f_ing", data = solet[solet$Muuttuja == "TEQ-imeytymisosuus" , ]["Result"])
f_mtoc <- Ovariable("f_mtoc", data = solet[solet$Muuttuja == "TEQ-osuus lapseen" , ]["Result"])
BF <- Ovariable("BF", data = solet[solet$Muuttuja == "Imeväisen rasvamäärä" , ]["Result"])
BF <- EvalOutput(BF) # LISÄTTY 13.3.
temp <- exposure
temp@output <- temp@output[temp@output$Exposure_agent == "TEQ" , ]
temp <- log((temp * t0.5 * f_ing * f_mtoc / (log(2) * BF) ) + 1) # Actual conversion
temp@output$Exposure_agent <- "logTEQ"
temp <- unkeep(temp, prevresults = TRUE, sources = TRUE)
exposure@output <- orbind(exposure, temp)
exposure <- unkeep(exposure, prevresults = TRUE, sources = TRUE)
frexposed <- 1
bgexposure <- addexposure
######################################################### VÄESTÖ
# Tarkastellaan totcases-laskennassa aluksi yksilöriskiä, ja vasta lopussa kerrotaan yksilötulokset yksilöiden edustamien ryhmien koolla.
population <- Ovariable(
"population",
data = data.frame(
Country = c("DK", "EST", "FI", "SWE"),
Result = c(2400000, 500000, 2700000, 4000000)
)
)
# Väkimäärä. TÄTÄ KÄYTETÄÄN disincidence-ovariablen suhteuttamisessa CHD:n suhteen.
######################## NOT SURE THIS IS THE BEST WAY
#!!+++++++++++++++++++++++++++++++++++++++++++++++++++++++++++
pop <- opbase.data("Op_en2949") # [[Population of Finland]]
#ii+++++++++++++++++++++++++++++++++++++++++++++++++++++++++++
pop$Obs <- NULL
pop$Observation <- NULL
age <- pop$Result[pop$Age == "0-4"]
pop$Result[pop$Age == "0-4"] <- 4/5 * age
pop <- orbind(pop, data.frame(Age = "0", Result = 1/5 * age))
levels(pop$Age)[levels(pop$Age) == "0-4"] <- "1-4"
pop <- Ovariable("pop", data = pop)
levels(pop@data$Age)
#### ADD sumitem HERE
#"1-4" "10-14" "15-19" "20-24" "25-29" "30-34" "35-39" "40-44" "45-49" "5-9"
#[11] "50-54" "55-59" "60-64" "65-69" "70-74" "75-79" "80-" "All" "0"
###############################################################################################
# ANNOSVASTEET JA TAUTIRISKIT
# dioksiinille,
# omega-3-rasvahapoille,
# vitamiineille (D-vitamiinille)
# ja metyylielohopealle ja niiden vaikutuksille
# Kuitenkaan metyylielohopeapitoisuuksia ei ole tässä malliversiossa, joten ne tippuvat pois.
# Annosvasteet
#!!+++++++++++++++++++++++ THIS CODE REPLACES ALL POLLUTANT-SPECIFIC CODES:
objects.latest("Op_en2031", code_name = "initiate") # [[Exposure-response function]]
# Drop ERF rows that are not used in this assessment.
#################### NOT SURE THERE IS REASON TO REMOVE
erfpois <- paste(ERF@data$Exposure_agent, ERF@data$Trait, ERF@data$ERF_parameter, ERF@data$Scaling) %in%
c(
"MeHg Child's IQ ERS BW",
"DHA Child's intelligence ERS BW",
"Omega3 Coronary heart disease RR None",
"Omega3 CHD Relative Hill None",
"logTEQ Developmental dental defects incl. agenesis ERS None",
"TEQ Cancer, total CSF BW",
"logTEQ Tooth defect ERS None" # Poistetaan Seveso koska suomalainen ERF uskottavampi
)
ERF@data <- ERF@data[!erfpois , ]
# Drop nuisance indices because they use a lot of memory in oapply.
################### SHOULD NOT USE A LOT OF MEMORY WITH aggregate-based oapply.
ERF@output <- ERF@output[ERF@output$Age != "<14" , ]
threshold@output <- threshold@output[threshold@output$Age != "<14" , ]
dropp <- c("Age", "Response_metric", "Exposure_route", "Exposure_metric", "Exposure_unit")
ERF <- unkeep(EvalOutput(ERF), dropp, sources = TRUE)
threshold <- unkeep(EvalOutput(threshold), dropp, sources = TRUE)
######################################## Tautiriski
################################### MAKE A SINGLE DISINCIDENCE OVARIABLE FROM THE CODE BELOF UNTIL HIA.
#!!++++++++++++++++++++++++++++++++++++++++++++++++++
objects.latest("Op_en5917", code_name = "initiate") # [[:op_en:Disease risk]] ovariable disincidence
#ii++++++++++++++++++++++++++++++++++++++++++++++++++
disincidence <- EvalOutput(disincidence) # TEMPORARY 13.3.
disincidence <- disincidence / 100000 # Change from 1/100000py to 1/py.
disincidence <- unkeep(disincidence, cols = c("Age", "Sex", "Population", "Unit", "Response_metric"))
# Näitä indeksejä ei tarvita tässä arvioinnissa.
disincidence@output <- disincidence@output[disincidence@output$Response != "CHD" , ] # Tulee ikävakioidusta alta.
############################################################################
# Ikävakioidut kuolemansyyt
# Sairastuvuus (tarkemmin sanottuna [[Kuolemansyyt Suomessa]])
#!!++++++++++++++++++++++++++++++++++++++++++++++++++++
syy <- Ovariable(
"syy",
data = opbase.data(
"Op_fi4558",
subset = "2012",
include = list(Kuolemansyy = c(
"04-21 Syövät (C00-C97)",
"27 Iskeemiset sydäntaudit (I20-I25)",
"29 Aivoverisuonien sairaudet (I60-I69)"
)
)
)
)
#ii++++++++++++++++++++++++++++++++++++++++++++++++++++
syy@data$Ikä <- gsub(" ", "", syy@data$Ikä)
colnames(syy@data)[colnames(syy@data) == "Ikä"] <- "Age"
syy@data <- syy@data[syy@data$Sukupuoli != "Sukupuolet yhteensä" , ]
# Syöpää ei haluta linkata ikäryhmittäin ERF:iin, vaan sitä käytetään muodostamaan diskontattu elinaikainen
# odotettu eliniän menetys jos kuolee syöpään tietyn ikäisenä. Ks. myöhemmin.
väli <- Ovariable(output = data.frame(
Kuolemansyy = c(
rep("27 Iskeemiset sydäntaudit (I20-I25)", 2),
"04-21 Syövät (C00-C97)",
"29 Aivoverisuonien sairaudet (I60-I69)"
),
Response = c("CHD", "CHD2", "Syövät", "Stroke"), # Syövät nimetään hassusti, koska sen EI haluta linkkautuvan ERF:iin.
# Lisäksi Tehdään kaksinkertainen sydäntaululistaus, koska on kaksi ERFiä: Mozaffarian ja Cohen.
Result = 1
))
## These rows should not be needed:
pop <- CollapseMarginal(EvalOutput(pop), cols = "Age", fun = "sum")
syy <- CollapseMarginal(EvalOutput(syy), cols = "Age", fun = "sum")
syyt <- syy / (pop / 2) * väli # Pop ei ole sukupuolen mukaan jaoteltu mutta syy on, joten pistetään kahtia.
disincidence <- disincidence * Ovariable(output = data.frame(Result = 1), marginal = FALSE)
marginals <- union(colnames(syyt@output)[syyt@marginal], colnames(disincidence@output)[disincidence@marginal])
syyt@output <- orbind(disincidence, syyt)
syyt@marginal <- colnames(syyt@output) %in% marginals
syyt <- unkeep(syyt, cols = c("Kuolemansyy", "Observation"), prevresults = TRUE, sources = TRUE)
colnames(syyt@output)[colnames(syyt@output) == "Sukupuoli"] <- "Gender"
syyt@output$Gender[syyt@output$Gender %in% c("Mies", "Miehet")] <- "Male" # Ei ole faktori vaan character
syyt@output$Gender[syyt@output$Gender %in% c("Nainen", "Naiset")] <- "Female"
syyt@output <- fillna(syyt@output, "Gender")
disincidence <- syyt # Nyt sisältää sekä alkuperäisen disincidencen että ikäluokittaisen CHD:n
###################################################################################
#### HIA-ovariablet
#!!++++++++++++++++++++++++++++++++++++++++++++++++++
objects.latest("Op_en2261", code_name = "totcases") # [[:op_en:Health impact assessment]]
# ovariable totcases and recursive ovariables dose, RR, and threshold
#ii++++++++++++++++++++++++++++++++++++++++++++++++++
totcases <- EvalOutput(totcases)
# WHY ARE THERE RESPONSES THAT DO NOT MATCH WITH EXPOSURES?
ggplot(totcases@output, aes(x = Response, weight = totcasesResult, fill = Gender))+
geom_bar()+coord_flip() + theme_gray(base_size = 24)
ggplot(exposure@output, aes(x = Result, colour = Gender))+
geom_density() + scale_x_log10() + theme_gray(base_size = 24)+
facet_wrap(~ Country)+labs(title = "Dioxin exposure (pg/d)")
colnames(exposure@output)
temp <- oapply(totcases, cols = "Iter", FUN = mean)
oprint(temp@output)
Loctable <- showLoctable()
oprint(showLoctable())
oprint(Loctable[Loctable$Index == "Age",])
##################################################################################
# Tapauskohtaiset postprosessoinnit
if(FALSE) {
### Muutetaan exposure yksikköön g /d kaikkien Exposure_agentien osalta.
skaala <- Ovariable(
output = data.frame(
Exposure_unit = "g /d",
Exposure_agent = c("Vitamin_D", "EPA", "DHA", "Omega3", "PCDDF", "PCB", "TEQ", "MeHg"),
Result = c(1E-6, 1E-3, 1E-3, 1E-3, 1E-12, 1E-12, 1E-12, 1E-6)),
marginal = c(FALSE, TRUE, FALSE)
)
tiedot <- rivit * background * Ovariable(
output = data.frame(
silakka[c("Rivi", "Sukupuoli", "Age", "Hedelm", "Ikä", "Paino", "Silakkamäärä")],
Result = 1
),
marginal = c(FALSE, TRUE, FALSE, TRUE, FALSE, FALSE, FALSE, FALSE)
)
tiedot <- unkeep(tiedot, cols = "Rivi", prevresults = TRUE, sources = TRUE)
exposure <- exposure * skaala * tiedot
amount <- amount * tiedot
concentration <- unkeep(concentration, prevresults = TRUE, sources = TRUE) * skaala
#Etsitään hedelmällisessä iässä olevat naiset ja lasten ÄO-vaste ja korjataan synnytyksen todennäköisyydellä.
indivrisk <- totcases
indivrisk <- indivrisk * tiedot
result(indivrisk) <- result(indivrisk) * ifelse(
indivrisk@output$Trait %in% c("Child's IQ", "Tooth defect", "Dental defect"),
ifelse(
indivrisk@output$Sukupuoli == "Nainen" & indivrisk@output$Hedelm == "TRUE",
0.1, # Probability of birth during a year.
0
),
1
)
#ages <- c(
# "0", "1-4", "5-9", "10-14", "15-19", "20-24",
# "25-29", "30-34", "35-39", "40-44", "45-49", "50-54",
# "55-59", "60-64", "65-69", "70-74", "75-79"
#)
indivrisk@output$Age <- factor(indivrisk@output$Age, levels = ages, ordered = TRUE)
indivrisk@output$Iter <- as.numeric(as.character(indivrisk@output$Iter))
#!!+++++++++++++++++++++++++++++++++++++++++++++++++
# objects.store(indivrisk, pop, exposure, amount, ERF, threshold, silakka, disincidence, concentration, rivit)
#ii+++++++++++++++++++++++++++++++++++++++++++++++++
cat("indivrisk, pop, exposure, amount, ERF, threshold, silakka, disincidence, concentration, rivit stored. \n")
####################################3
for(i in c("RR", "disincidence", "totcases", "ERF", "threshold")) {
print(levels(get(i)@output$Response))
}
} # if(FALSE)
######################################## TESTIALUE
totcases2 <- Ovariable("totcases", # This calculates the total number of cases in each population subgroup.
# The cases are calculated for specific (combinations of) causes. However, these causes are NOT visible in the result.
dependencies = data.frame(
Name = c(
"population", # Population divided into subgroups as necessary
"dose", # Exposure to the pollutants
"disincidence", # Incidence of the disease of interest
"RR", # Relative risks for the given exposure
"ERF", # Other ERFs than those that are relative to background.
"threshold", # exposure level below which the agent has no impact.
"frexposed" # fraction of population that is exposed
),
Ident = c(
"Op_en2261/population", # [[Health impact assessment]]
"Op_en2261/dose", # [[Health impact assessment]]
"Op_en5917/initiate", # [[Disease risk]]
"Op_en2261/RR", # [[Health impact assessment]]
"Op_en2031/initiate", # [[Exposure-response function]]
"Op_en2031/initiate", # [[Exposure-response function]]
"Op_en2261/frexposed" # [[Health impact assessment]]
)
),
formula = function(...) {
test <- list()
ERF@marginal[colnames(ERF@output) %in% c("ERF_parameter", "Scaling")] <- FALSE # Make sure that these are not marginals
############### First look at the relative risks based on RR
if(testforrow(RR, dose)) { # If an ovariable whose nrow(ova@output) == 0
# is used in Ops, it is re-EvalOutput'ed, and therefore ERFrr*dose may have rows even if ERFrr doesn't.
# takeout is a vector of column names of indices that ARE in population but NOT in the disease incidence.
# However, populationSource is kept because oapply does not run if there are no indices.
if(FALSE) {
print("RR")
print(head(dose@output))
if(class(population) == "ovariable") {
takeout <- setdiff(colnames(population@output)[population@marginal],
colnames(disincidence@output)[disincidence@marginal]
)
if(length(takeout) > 0) {# Aggregate to larger subgroups.
pop <- oapply(population, NULL, sum, takeout)
} else {
pop <- population
}
} else {
takeout <- character()
pop <- population
}
# pci is the proportion of cases across different population subroups
# based on differential risks and
# population sizes. pci sums up to 1 for each larger subgroup found in disincidence.
# See [[Population attributable fraction]].
pci <- population * RR
print("pci1")
print(head(pci@output))
# Divide pci by the values of the actually exposed group (discard nonexposed)
# The strange Ovariable thing is needed to change the name of temp to avoid problems later.
temp <- pci * Ovariable(data = data.frame(Result = 1))
if ("Exposcen" %in% colnames(temp@output)) {
temp@output <- temp@output[temp@output$Exposcen == "BAU" , ]
temp <- unkeep(temp, cols = "Exposcen", prevresults = TRUE, sources = TRUE)
}
temp <- unkeep(temp, prevresults = TRUE, sources = TRUE)
if(length(takeout) > 0) temp <- oapply(temp, NULL, sum, takeout)
#if(length(takeout) > 0) temp <- ooapply(temp, cols = takeout, FUN = "sum", use_plyr = TRUE)
# if(length(takeout) > 0) temp <- osum(temp, cols = takeout)
print("temp")
print(head(temp@output))
pci <- pci / temp
temp <- NULL
} # if(FALSE)
# The cases are divided into smaller subgroups based on weights in pci.
# This is why the larger groups of population are used (pop instead of population).
out1 <- disincidence * population * (RR-1)/RR #unkeep(pci, prevresults = TRUE, sources = TRUE)
# out1 <- unkeep(out1, cols = "populationResult") # populationResult comes from pop and not from pci that actually contains
# the population weighting for takeout indices. Therefore it would be confusing to leave it there.
#print("out1")
#print(head(out1@output))
test <- c(test, out1)
}
out1 <- NULL
##########################################################################
############# This part is about absolute risks (i.e., risk is not affected by background rates).
# Unit risk (UR), cancer slope factor (CSF), and Exposure-response slope (ERS) estimates.
UR <- ERF
UR@output <- UR@output[UR@output$ERF_parameter %in% c("UR", "CSF", "ERS") , ]
if(testforrow(UR, dose)) { # See RR for explanation.
UR <- threshold + UR * dose * frexposed # Actual equation
# threshold is here interpreted as the baseline response (intercept of the line). It should be 0 for
# UR and CSF but it may have meaningful values with ERS
print("UR")
print(head(dose@output))
UR <- oapply(UR, NULL, sum, "Exposure_agent")
UR <- population * UR
test <- c(test, UR)
}
UR <- NULL
# Step estimates: value is 1 below threshold and above ERF, and 0 in between.
# frexposed cannot be used with Step because this may be used at individual and maybe at population level.
Step <- ERF
Step@output <- Step@output[Step@output$ERF_parameter %in% c("Step", "ADI", "TDI", "RDI", "NOAEL") , ]
if(testforrow(Step, dose)) { # See RR for explanation.
Step <- 1 - (dose >= threshold) * (dose <= Step) # Actual equation
# Population size should be taken into account here. Otherwise different population indices may go unnoticed.(?)
Step <- oapply(Step, NULL, sum, "Exposure_agent")
test <- c(test, Step)
}
Step <- NULL
#####################################################################
# Combining effects
if(length(test) == 0) return(data.frame())
if(length(test) == 1) out <- test[[1]]@output
if(length(test) == 2) out <- orbind(test[[1]], test[[2]])
if(length(test) == 3) out <- orbind(orbind(test[[1]], test[[2]]), test[[3]])
# Find out the right marginals for the output
marginals <- character()
nonmarginals <- character()
for(i in 1:length(test)) {
marginals <- c(marginals, colnames(test[[i]]@output)[test[[i]]@marginal])
nonmarginals <- c(nonmarginals, colnames(test[[i]]@output)[!test[[i]]@marginal])
}
test <- NULL
out <- out[!colnames(out) %in% c("populationSource", "populationResult")] # These are no longer needed.
out <- Ovariable(output = out, marginal = colnames(out) %in% setdiff(marginals, nonmarginals))
if("Exposcen" %in% colnames(out@output)) {
out <- out * Ovariable(
output = data.frame(Exposcen = c("BAU", "No exposure"), Result = c(1, -1)),
marginal = c(TRUE, FALSE)
)
out <- oapply(out, NULL, sum, "Exposcen")
}
return(out)
}
)
totcases2 <- EvalOutput(totcases2)
unique(totcases2@output$Response)
nrow(RR@output)
oprint(head(totcases2@output))
ggplot(totcases2@output, aes(x = Response, weight = totcasesResult, fill = Gender))+geom_bar(position="dodge")
| |