Insight network: Difference between revisions
mNo edit summary |
|||
| (33 intermediate revisions by the same user not shown) | |||
| Line 36: | Line 36: | ||
gr <- makeInsightGraph(graphTable) # Creates a DiagrammeR graph object | gr <- makeInsightGraph(graphTable) # Creates a DiagrammeR graph object | ||
shinyApp(ui, server) | shinyApp(ui, server) | ||
| Line 49: | Line 47: | ||
=== Process === | === Process === | ||
:''Insight networks have been described in a scientific article manuscript [[From open assessment to shared understanding: practical experiences#Insight networks]]. Objects and their relations used in [[open policy practice]] are described on page [[Open policy ontology]]. | :''Insight networks have been described in a scientific article manuscript [[From open assessment to shared understanding: practical experiences#Insight networks]]. Objects and their relations used in [[open policy practice]] are described on page [[Open policy ontology]]. | ||
| Line 192: | Line 53: | ||
These diagrams use graph theory with vertices (or nodes) and arcs (or arrows). They are used to describe and define all the pieces needed for a description of the situation under scrutiny. Diagrams may be produced with any graphics software, providing that calculation functions are not required. If calculations ''are'' needed, we recommend the use of [[R]] software and [[OpasnetUtils]] package. | These diagrams use graph theory with vertices (or nodes) and arcs (or arrows). They are used to describe and define all the pieces needed for a description of the situation under scrutiny. Diagrams may be produced with any graphics software, providing that calculation functions are not required. If calculations ''are'' needed, we recommend the use of [[R]] software and [[OpasnetUtils]] package. | ||
This is the process how data flows into insight diagrams: | This is the process how data flows into insight diagrams: | ||
| Line 254: | Line 68: | ||
In the next phase, each csv file is opened, interpreted, and defined as items and relations. This is done in code Op_fi5810/graphs on page [[:op_fi:Ympäristöterveysindikaattori]]. All these are saved as a DiagrammeR graph, and each topic may be separately selected as a subgraph. | In the next phase, each csv file is opened, interpreted, and defined as items and relations. This is done in code Op_fi5810/graphs on page [[:op_fi:Ympäristöterveysindikaattori]]. All these are saved as a DiagrammeR graph, and each topic may be separately selected as a subgraph. | ||
* {{argument|relat1=relevant attack|truth1=true|id=0094|type=|content=Are Tehtäväkokonaisuus, Osiotyyppi, JHS-luokka actually types of objects, or are they just indices. Yes, they should be indices and the objects relate to them with "has index". Correct table 4.|sign=--[[User:Jouni|Jouni]] ([[User talk:Jouni|talk]]) 20:46, 17 July 2018 (UTC)}} | * {{argument|relat1=relevant attack|truth1=true|id=0094|type=|content=Are Tehtäväkokonaisuus, Osiotyyppi, JHS-luokka actually types of objects, or are they just indices. Yes, they should be indices and the objects relate to them with "has index". Correct table 4.|sign=--[[User:Jouni|Jouni]] ([[User talk:Jouni|talk]]) 20:46, 17 July 2018 (UTC)}} | ||
* {{argument|relat1=relevant attack|truth1=true|id=0095|type=|content=Check the code about renderging graphs and creating a server function that also works on non-web environments on [[:op_fi:Keskustelu:Näkemysverkko#Näkemysverkkoabstrakti vaikuttavuuden tutkimuksen päiville 4.-5.12.2018]]|sign=--[[User:Jouni|Jouni]] ([[User talk:Jouni|talk]]) 20:46, 17 July 2018 (UTC)}} | |||
=== Data === | === Data === | ||
| Line 261: | Line 76: | ||
<t2b name="Graphical properties of objects and relations" index="Property,Value,Parameter" obs="Result" desc="Description" unit="-"> | <t2b name="Graphical properties of objects and relations" index="Property,Value,Parameter" obs="Result" desc="Description" unit="-"> | ||
default|default|node.shape|circle|Default values unless something else is specified | default|default|node.shape|circle|Default values unless something else is specified | ||
default|default|node.style|filled| | |||
default|default|node.sides|4| | default|default|node.sides|4| | ||
default|default|node.skew|0| | default|default|node.skew|0| | ||
| Line 303: | Line 119: | ||
type|process|node.shape|pentagon|Process type object | type|process|node.shape|pentagon|Process type object | ||
type|process|node.fillcolor|purple1|Process type object | type|process|node.fillcolor|purple1|Process type object | ||
type|action|node.fillcolor| | type|action|node.fillcolor|#009246|Process type object, dark green (0,146,70) | ||
type|action|node.shape|rectangle|Decision type object | |||
type|task 1|node.color|brown|Illustration of the responsible organisation of the task | type|task 1|node.color|brown|Illustration of the responsible organisation of the task | ||
type|task 2|node.color|yellow|Illustration of the responsible organisation of the task | type|task 2|node.color|yellow|Illustration of the responsible organisation of the task | ||
| Line 309: | Line 126: | ||
type|task 4|node.color|green|Illustration of the responsible organisation of the task | type|task 4|node.color|green|Illustration of the responsible organisation of the task | ||
type|task 5|node.color|red|Illustration of the responsible organisation of the task | type|task 5|node.color|red|Illustration of the responsible organisation of the task | ||
type|decision|node.fillcolor|red|Decision type object | type|decision|node.fillcolor|red|Decision type object | ||
type|data|node.shape|rectangle|Data type object | type|data|node.shape|rectangle|Data type object | ||
| Line 323: | Line 139: | ||
type|true statement|node.fillcolor|gold|Argument type object | type|true statement|node.fillcolor|gold|Argument type object | ||
type|false statement|node.fillcolor|gray|Argument type object | type|false statement|node.fillcolor|gray|Argument type object | ||
type|fact opening statement|node.fillcolor|lightskyblue1|Argument type object | type|fact opening statement|node.fillcolor|lightskyblue1|Argument type object. Discussion start | ||
type|value opening statement|node.fillcolor|palegreen1|Argument type object | type|value opening statement|node.fillcolor|palegreen1|Argument type object | ||
type|fact closing statement|node.fillcolor|skyblue|Argument type object | type|fact closing statement|node.fillcolor|skyblue|Argument type object. Discussion end | ||
type|value closing statement|node.fillcolor|springgreen|Argument type object | type|value closing statement|node.fillcolor|springgreen|Argument type object. | ||
type|fact discussion|node.fillcolor|skyblue|Argument type object. Not neede? | type|fact discussion|node.fillcolor|skyblue|Argument type object. Not neede? | ||
type|value discussion|node.fillcolor|springgreen|Value judgement type object. Not needed? | type|value discussion|node.fillcolor|springgreen|Value judgement type object. Not needed? | ||
| Line 332: | Line 148: | ||
type|indicator|node.color|brown|Additional information about object class | type|indicator|node.color|brown|Additional information about object class | ||
type|indicator|node.fillcolor|gold|Additional information about object class | type|indicator|node.fillcolor|gold|Additional information about object class | ||
type|operational indicator|node.fillcolor|#00d7a7|Additional information about object class light green (0,215,167) | |||
type|tactical indicator|node.fillcolor|#9fc9eb|Additional information about object class light blue (159,201,235) | |||
type|strategic indicator|node.fillcolor|#0072c6|Additional information about object class dark blue (0,114,198) | |||
type|strategic indicator|node.shape|diamond|Additional information about object class | |||
type|arviointikriteeri|node.color|orange|Not quite clear what criteria objects are: indicators or value statements, or something else | type|arviointikriteeri|node.color|orange|Not quite clear what criteria objects are: indicators or value statements, or something else | ||
type|task|node.color|green|Additional information about object class | type|task|node.color|green|Additional information about object class | ||
| Line 338: | Line 158: | ||
Relation|causal link|edge.color|black|Causal link | Relation|causal link|edge.color|black|Causal link | ||
Relation|causal link|edge.style|solid|Causal link | Relation|causal link|edge.style|solid|Causal link | ||
Relation|positive causal link|edge.fontcolor|green|Causal link | Relation|positive causal link|edge.fontcolor|#009246|Causal link, dark green (0,146,70) | ||
Relation|negative causal link|edge.fontcolor|red|Causal link | Relation|increases|edge.fontcolor|#009246|Causal link, dark green (0,146,70) | ||
Relation|negative causal link|edge.fontcolor|#bd2719|Causal link, red (189,39,25) | |||
Relation|decreases|edge.fontcolor|#bd2719|Causal link, red (189,39,25) | |||
Relation|part_of|edge.fontcolor|gray|Part of (set theory link) | |||
Relation|participatory link|edge.color|purple|Participatory link | Relation|participatory link|edge.color|purple|Participatory link | ||
Relation|participatory link|edge.style|dashed|Participatory link | Relation|participatory link|edge.style|dashed|Participatory link | ||
| Line 355: | Line 178: | ||
</t2b> | </t2b> | ||
==== | === Calculations === | ||
'''Insight network 2.0 | |||
An updated version should improve the | |||
* a) context sensitivity (referring to primarily to objects within own context but secondarily to those from another context), | |||
* b) making graphs by default from a single context rather than a full list of contexts from a meta table, | |||
* c) compatibility with cytoscape.js, | |||
* d) merging ready-made graphs meaningfully, | |||
* e) have a reasonable intermediate object format that contains all data needed, such as | |||
** tables for nodes and edges, compatible with Diagrammer, Cytoscape.js, AND Gephi. | |||
** metadata for display, such as seeds, steps, object types to ignore, whether to show labels etc. Or should these just be implemented on the graph? | |||
What should be done? | |||
# Fetch the data table by scrape or other function and with data about URL, table, and initial row. | |||
# Use splizzeria and fillprev if needed. | |||
# Interpret columns based on a vector of column numbers (with possibly 1+2 notation to paste columns) to create the standard columns. If this is done in an ovariable formula, there is no need for a specific function. | |||
#* Context | |||
#* Item | |||
#* type | |||
#* label | |||
#* rel | |||
#* Object | |||
#* Description | |||
#* Reldescription | |||
#* URL | |||
#* Result (dummy, always 0) | |||
# Create missing node rows from objects. Do NOT assume context. | |||
# Create URL from permanent resource location trunk and the identifier (where does the identifier come from?) | |||
# Item ja label laitetaan pötköön ja haetaan mätsi. Tulos onrow-pötköstä. | |||
# Create an ovariable from the table. | |||
# Add meta to the ovariable with formatting data. | |||
#* insightGraph: | |||
#** seed | |||
#** removenodes | |||
#** formatting (character vector with possible entries: Hide node labels, Hide edge labels, Show legend nodes, Remove branches only) | |||
#** ignoreobj | |||
#** steps | |||
# (NOT NEEDED? Create Oldid if does not exist from context and numbering) | |||
# If a relation is presented as item, the formatting is applied to the ring. | |||
Combine graph objects | |||
* Find items without context. Match them with items with the same Item (label) that do have a type. | |||
Tuplarelaatiot, voidaanko kategorisesti poistaa? | |||
Out <- rep(NA, length(find)) | |||
For(x in cond,) | |||
For(i in 1:length(find) | |||
Tmp<-id[context==contextfind(i))])[Match(find(i), df$cond(x)(df$context==contextfind(i))] pitää etsiä id alkuperäisestä taulukosta heti muuten ei toimi | |||
Out<- ifelse(isna(out). Tmp,out) | |||
)) | |||
Sitten sama ioman contekstirajoitusta. | |||
'''Insight network 1.0 | |||
There are three different identifiers for a subject item. | |||
* Oldid: a technical identifier typically of format context.number, where number is a sequential number within a context. | |||
* Item: the actual name of the item, detailed enough to give a good understanding of its meaning. | |||
* label: a short name shown on insight networks. Does not exmplain everything, just enough to distinguish it from other items. | |||
If Oldid is not given, it is created from the context and a number. If label is not given in data, it is truncated from Item. | |||
Object item has one column ''Object'' that may contain any of these. The priority is Item > label > Oldid > Object. The last option means that it is assumed that Object refers to a new item that is not mentioned in the Item column. | |||
An insight network is produced in this order (last object mentioned first). | |||
# | # gr: a diagrammer graph with all data and formatting for an insight network. Produced by makeInsightGraph. | ||
# makeInsightGraph | |||
==== Making insight graphs ==== | |||
<rcode name="formatted" label="Initiate data.frame formatted (for developers only)" embed=1> | |||
# This is code Op_en3861/formatted on page [[Insight network]] | |||
library(OpasnetUtils) | library(OpasnetUtils) | ||
#' | #' Function formatting creates a formatting table for nodes and edges. | ||
#' | #' The function as no parameters. | ||
#' | #' @return data.frame with ontology terms as rows and formatting properties as columns. | ||
# | |||
#' | formatting <- function() { | ||
# | ## Replace default setting with additional class info | ||
# | |||
## Find all classes for item subclasses | |||
hierItem <- opbase.data("Op_en7783", subset="Item types") # [[Open policy ontology]] | |||
hierItem <- hierItem[c("Object","English name","Finnish name")] | |||
colnames(hierItem)[colnames(hierItem)=="Object"] <- "Class" # Contains also other relations than subclass, notably "part of". | |||
## Find all classes for relation subclasses | |||
hierRel <- opbase.data("Op_en7783", subset="Relation types") # [[Open policy ontology]]. All relations are of type 'has subclass' | |||
for(i in colnames(hierRel)) hierRel[[i]] <- as.character(hierRel[[i]]) | |||
hierRel <- data.frame( | |||
Class = rep(hierRel$Class, 2), | |||
Tmp1 = c(hierRel$`English name`, hierRel$`English inverse`), | |||
Tmp2 = c(hierRel$`Finnish name`, hierRel$`Finnish inverse`) | |||
) | |||
colnames(hierRel) <- c("Class", "English name", "Finnish name") | |||
# Make a single resource list | |||
hier <- rbind(hierItem, hierRel) | |||
for(i in colnames(hier)) hier[[i]] <- as.character(hier[[i]]) | |||
# Combine language versions of resource list | |||
hier <- unique(data.frame( | |||
Class = c(hier$Class, rep(hier$`English name`, 2)), | |||
Item = c(rep(hier$`English name`, 2), hier$`Finnish name`), | |||
stringsAsFactors = FALSE | |||
)) | |||
out <- hier | |||
tmp <- out$Class | |||
for(i in 1:6) { | |||
tmp <- hier$Class[match(tmp, hier$Item)] | |||
out <- rbind( | |||
out, | |||
cbind( | |||
Class = tmp, | |||
Item = hier$Item | |||
) | |||
) | |||
} | |||
hier <- out[!is.na(out$Class),] | |||
# Replace default settings with property-specific settings | |||
# First fetch the graphical styles of properties from [[Insight network]] | |||
prop_gen <- opbase.data( | |||
"Op_en3861", # [[Insight network]] | |||
subset="Graphical properties of objects and relations" | |||
) | |||
tmp <- prop_gen[grepl("edge", prop_gen$Parameter) & prop_gen$Value!="default",] | |||
tmp$Parameter <- gsub("edge","node",tmp$Parameter) | |||
prop_gen <- rbind(prop_gen, tmp) | |||
# Create a data.frame with all item * parameter combinations. | |||
# This will be filled with item-specific graph settings | |||
for(i in | |||
prop_spec <- merge( | |||
data.frame(Resource = unique(c(hier$Class, hier$Item))), | |||
prop_gen[prop_gen$Property=="default",c("Parameter","Result")] | |||
) | |||
for(i in 1:nrow(prop_gen)) { | |||
if(prop_gen$Property[i] != "default") { | |||
# Names of items that should have the property replaced | |||
tst <- unique(hier$Item[hier$Class==prop_gen$Value[i]]) | |||
prop_spec$Result[prop_spec$Resource %in% tst & prop_spec$Parameter==prop_gen$Parameter[i]] <- prop_gen$Result[i] | |||
} | } | ||
} | } | ||
return( | |||
prop_spec$Result <- as.character(prop_spec$Result) | |||
formatted <- reshape(prop_spec, idvar="Resource", timevar="Parameter", direction="wide") | |||
colnames(formatted) <- gsub("Result.", "", colnames(formatted)) | |||
# > colnames(formatted) | |||
# [1] "Resource" "node.shape" "node.sides" "node.skew" | |||
# [5] "node.fillcolor" "node.fontsize" "node.height" "node.width" | |||
# [9] "node.color" "node.penwidth" "node.fontcolor" "node.distortion" | |||
# [13] "edge.color" "edge.fontsize" "edge.fontcolor" "edge.style" | |||
# [17] "edge.penwidth" "edge.arrowsize" | |||
for(i in c( | |||
"node.sides", | |||
"node.skew", | |||
"node.fontsize", | |||
"node.height", | |||
"node.width", | |||
"node.penwidth", | |||
"node.distortion", | |||
"edge.fontsize", | |||
"edge.penwidth", | |||
"edge.arrowsize" | |||
)) formatted[[i]] <- as.numeric(formatted[[i]]) | |||
return(formatted) | |||
} | } | ||
objects.store( | formatted <- formatting() | ||
cat(" | |||
objects.store(formatted) | |||
cat("Data.frame formatted stored.\n") | |||
</rcode> | </rcode> | ||
<rcode name=" | <rcode name="makeGraph2" label="Initiate function makeGraph (for developers only)"> | ||
# This is code Op_en3861/ | # This is code Op_en3861/makeGraph2 on page [[Insight network]] | ||
library(OpasnetUtils) | library(OpasnetUtils) | ||
#' | #' Making insight network graph object | ||
#' | #' | ||
#' | #' makeGraph is a function for taking an insight ovariable and making a graph object. | ||
#' | #' | ||
#' @param | #' @param a is data.frame defining nodes and edges with at least columns: Oldid, type, Item, label, Relation, Object, Description. Other columns for nodes such as URL are allowed. | ||
#' @return two data.frames: nodes_df and edges_df that are directly given as parameters for DiagrammeR::create_graph. | |||
#' @return data. | |||
makeGraph <- function(ova, formatting=data.frame(), ...) { | |||
require(OpasnetUtils) | |||
require(DiagrammeR) | |||
if(!exists("formatted") & nrow(formatted)==0){ | |||
objects.latest("Op_en3861", code_name="formatted") # [[Insight network]] formatted | |||
} | |||
if(!exists("chooseGr")) { | |||
objects.latest("Op_en3861", code_name="chooseGr") # [[Insight network]] chooseGr | |||
} | |||
if("ovariable" %in% class(ova)) { | |||
a <- ova@output | |||
meta <- ova@meta$insightnetwork | |||
} else { | |||
a <- ova | |||
meta <- NULL | |||
} | |||
a$truth <- signif(a$truth,2) | |||
a$relevance <- signif(a$relevance,2) | |||
for(i in 1:ncol(a)) { | |||
a[[i]] <- gsub("[\"']", " ", a[[i]]) | |||
} | |||
# Fill in missing labels, Items, and object nodes | |||
a$label <- ifelse(is.na(a$label),substr(a$Item,1,30), a$label) | |||
a$Item <- ifelse(is.na(a$Item),a$label, a$Item) | |||
# Find nrow that matches the Object based on Item or label. | |||
tst <- rep(1:nrow(a),2)[match(a$Object, c(a$Item, a$label))] | |||
# Use Item as Object identifier when possible | |||
hasobj <- !(is.na(a$Object) | a$Object=="") # Rows of data.frame a that have Object | |||
a$Object[hasobj] <- a$Item[tst][hasobj] | |||
# Find objects that have not been defined | |||
newbies <- ifelse(is.na(tst), a$Object,NA) | |||
newbies <- newbies[!is.na(newbies)] | |||
if(length(newbies)>0) { | |||
a <- orbind( | |||
a, | |||
data.frame( | |||
Item=newbies, | |||
label=substr(newbies,1,30), | |||
stringsAsFactors = FALSE | |||
) | |||
) | |||
} | |||
nodes <- a[!(duplicated(a$Item) | is.na(a$Item) | a$Item==""),] | |||
# nodes$tooltip <- paste0( | |||
# nodes$label, ". ", | |||
# ifelse(nodes$label == nodes$Item, "", paste0(nodes$Item, ". ")), | |||
# ifelse(is.na(nodes$Description), "", paste0("\n", nodes$Description)), | |||
# " (", nodes$Context, "/", nodes$id,")", | |||
# ) | |||
nodes$tooltip <- paste0( | |||
nodes$Item, ". ", nodes$Description, "/ truth: ", nodes$truth, " relevance: ", nodes$relevance) | |||
nodes <- merge(nodes, formatted[setdiff(colnames(formatted),colnames(nodes))], | |||
by.x="type", by.y="Resource") | |||
colnames(nodes) <- gsub("node.","",colnames(nodes)) | |||
nodes <- nodes[!grepl("edge.", colnames(nodes))] | |||
nodes$id <- 1:nrow(nodes) | |||
# Create edges and flip unpreferred relations to their inverse relations | |||
inver <- opbase.data("Op_en7783", subset="Relation types") | |||
for(i in colnames(inver)) inver[[i]] <- as.character(inver[[i]]) | |||
inve <- data.frame( | |||
rel = c(inver$`English name`,inver$`Finnish name`), | |||
inve = c(inver$`English inverse`,inver$`Finnish inverse`), | |||
stringsAsFactors = FALSE | |||
) | |||
edges <- a[!(is.na(a$Object) | a$Object=="") , ] | |||
flip <- edges$rel %in% inve$inve | |||
tmp <- edges$Item | |||
edges$Item[flip] <- edges$Object[flip] | |||
edges$Object[flip] <- tmp[flip] | |||
edges$rel[flip] <- inve$rel[match(edges$rel, inve$inve)][flip] | |||
edges$from <- match(edges$Item, nodes$Item) | |||
edges$to <- match(edges$Object, nodes$Item) | |||
edges$label <- edges$rel | |||
edges <- merge(edges, formatted[setdiff(colnames(formatted),colnames(edges))], | |||
by.x="rel", by.y="Resource") | |||
colnames(edges) <- gsub("edge.","",colnames(edges)) | |||
edges <- edges[!grepl("node.", colnames(edges))] | |||
edges$id <- 1:nrow(edges) | |||
edges$labeltooltip <- paste0(edges$label, " (",edges$Context, "/",edges$id, ")") | |||
gr <- create_graph( | |||
nodes_df=nodes, | |||
edges_df=edges | |||
) | |||
if(!is.null(meta)) { | |||
gr <- chooseGr(gr, input=meta) | |||
} | |||
return(gr) | |||
} | } | ||
objects.store( | objects.store(makeGraph) | ||
cat("Function | cat("Function makeGraph stored.\n") | ||
</rcode> | </rcode> | ||
<rcode name=" | <rcode name="makeGraph" label="Initiate function makeGraph (old version)" embed=1> | ||
# This is code Op_en3861/ | # This is code Op_en3861/makeGraph on page [[Insight network]] | ||
library(OpasnetUtils) | library(OpasnetUtils) | ||
#' Making insight network graph object | #' Making insight network graph object | ||
#' | #' | ||
#' | #' makeGraph is a function for taking an insight ovariable and making a graph object. | ||
#' | #' | ||
#' @param a is data.frame defining nodes and edges with at least columns: Oldid, type, Item, label, Relation, Object, Description. Other columns for nodes such as URL are allowed. | #' @param a is data.frame defining nodes and edges with at least columns: Oldid, type, Item, label, Relation, Object, Description. Other columns for nodes such as URL are allowed. | ||
#' @return | #' @return two data.frames: nodes_df and edges_df that are directly given as parameters for DiagrammeR::create_graph. | ||
makeGraph <- function(ova, ...) { | |||
require(OpasnetUtils) | require(OpasnetUtils) | ||
require(DiagrammeR) | require(DiagrammeR) | ||
if(!exists("formatted")){ | |||
objects.latest("Op_en3861", code_name="formatted") # [[Insight network]] formatted | |||
} | |||
if(!exists("chooseGr")) { | |||
objects.latest("Op_en3861", code_name="chooseGr") # [[Insight network]] chooseGr | |||
} | } | ||
if("ovariable" %in% class(ova)) { | |||
a <- ova@output | |||
meta <- ova@meta$insightnetwork | |||
} else { | |||
a <- ova | |||
meta <- NULL | |||
} | |||
for(i in 1:ncol(a)) { | |||
a[[i]] <- gsub("[\"']", " ", a[[i]]) | |||
} | } | ||
# | # Fill in missing labels, Items, and object nodes | ||
a$label <- ifelse(is.na(a$label),substr(a$Item,1,30), a$label) | |||
a$Item <- ifelse(is.na(a$Item),a$label, a$Item) | |||
# | # Find nrow that matches the Object based on Item or label. | ||
tst <- rep(1:nrow(a),2)[match(a$Object, c(a$Item, a$label))] | |||
# Use Item as Object identifier when possible | |||
hasobj <- !(is.na(a$Object) | a$Object=="") # Rows of data.frame a that have Object | |||
a$Object[hasobj] <- a$Item[tst][hasobj] | |||
# Find | # Find objects that have not been defined | ||
newbies <- ifelse(is.na(tst), a$Object,NA) | |||
newbies <- newbies[!is.na(newbies)] | |||
if(length(newbies)>0) { | |||
a <- orbind( | |||
a, | |||
data.frame( | |||
Item=newbies, | |||
label=substr(newbies,1,30), | |||
stringsAsFactors = FALSE | |||
) | |||
) | ) | ||
} | |||
nodes <- | nodes <- a[!(duplicated(a$Item) | is.na(a$Item) | a$Item==""),] | ||
nodes$tooltip <- paste0( | nodes$tooltip <- paste0( | ||
nodes$label, ". ", | nodes$label, ". ", | ||
ifelse(nodes$label == nodes$Item, "", paste0(nodes$Item, ". ")), | ifelse(nodes$label == nodes$Item, "", paste0(nodes$Item, ". ")), | ||
ifelse(is.na(nodes$Description), "", paste0("\n", nodes$Description)), | ifelse(is.na(nodes$Description), "", paste0("\n", nodes$Description)), | ||
" (", nodes$Context, "/", nodes$id,")" | |||
" (", nodes$ | |||
) | ) | ||
nodes <- merge(nodes, formatted, by.x="type", by.y="Resource") | |||
colnames(nodes) <- gsub("node.","",colnames(nodes)) | |||
nodes <- nodes[!grepl("edge.", colnames(nodes))] | |||
nodes$id <- 1:nrow(nodes) | |||
# Create edges and flip unpreferred relations to their inverse relations | # Create edges and flip unpreferred relations to their inverse relations | ||
inver <- opbase.data("Op_en7783", subset=" | inver <- opbase.data("Op_en7783", subset="Relation types") | ||
for(i in colnames(inver)) inver[[i]] <- as.character(inver[[i]]) | |||
inve <- data.frame( | |||
rel = c(inver$`English name`,inver$`Finnish name`), | |||
inve = c(inver$`English inverse`,inver$`Finnish inverse`), | |||
stringsAsFactors = FALSE | |||
) | |||
edges <- a[!(is.na(a$Object) | a$Object=="") , ] | |||
edges <- a[!(is.na(a$Object) | a$Object=="") , | flip <- edges$rel %in% inve$inve | ||
flip <- edges$ | tmp <- edges$Item | ||
tmp <- edges$ | edges$Item[flip] <- edges$Object[flip] | ||
edges$ | |||
edges$Object[flip] <- tmp[flip] | edges$Object[flip] <- tmp[flip] | ||
edges$ | edges$rel[flip] <- inve$rel[match(edges$rel, inve$inve)][flip] | ||
edges$from <- match(edges$Item, nodes$Item) | |||
edges$to <- match(edges$Object, nodes$Item) | |||
edges$label <- edges$rel | |||
edges$labeltooltip <- paste0(edges$label, " (",edges$Context, "/",edges$id, ")") | |||
edges <- merge(edges, formatted, by.x="rel", by.y="Resource") | |||
colnames(edges) <- gsub("edge.","",colnames(edges)) | |||
edges <- edges[!grepl("node.", colnames(edges))] | |||
edges$id <- 1:nrow(edges) | edges$id <- 1:nrow(edges) | ||
gr <- create_graph( | |||
nodes_df=nodes, | |||
edges_df=edges | |||
) | |||
if(!is.null(meta)) { | |||
gr <- chooseGr(gr, input=meta) | |||
} | |||
return(gr) | |||
} | |||
objects.store(makeGraph) | |||
cat("Function makeGraph stored.\n") | |||
</rcode> | |||
<rcode name="chooseGr" label="Initiate ovariable chooseGr" embed=1> | |||
# This is code Op_en3861/chooseGr on page [[Insight diagram]]. | |||
library(OpasnetUtils) | |||
#' Function chooseGr takes a diagrammer graph and selects s subgraph based on topic, labels, steps from selected nodes etc. | |||
#' @param gr diagrammer graph | |||
#' @param input list of arguments to be used in selection | |||
#' @seeds ovariable where @data has columns Topic to be chosen and Node for Oldid's to select. | |||
#' @return diagrammer graph where node_selection contains the selected nodes | |||
chooseGr <- function(gr, input, seeds=NULL, verbose=FALSE) { | |||
if(!is.null(seeds)) seeds <- match(seeds@data$Node[seeds@data$Topic==input$topic], gr$nodes_df$Oldid) | |||
nods <- union(c( | |||
seeds, | |||
match(input$addnodes, gr$nodes_df$label)), | |||
match(input$addnodesByid, gr$nodes_df$id) | |||
) | ) | ||
nods <- nods[!is.na(nods)] | |||
gr <- deselect_nodes(gr, get_selection(gr)) | |||
for(i in 1:nrow( | gr <- select_nodes_by_id(gr, nods) | ||
if( | if(input$steps>0) { | ||
for(i in 1:input$steps) { | |||
gr <- deselect_nodes(gr,match(input$removenodes, gr$nodes_df$label)) | |||
if(nrow(gr$node_selection)>0) { | |||
gr <- trav_both(gr,add_to_selection = TRUE) | |||
} | |||
} | |||
if("Remove branches only" %in% input$formatting) { | |||
gr <- select_nodes_by_id(gr,match(input$removenodes, gr$nodes_df$label)) | |||
} else { | |||
gr <- deselect_nodes(gr,match(input$removenodes, gr$nodes_df$label)) | |||
} | } | ||
} | } | ||
if("Show legend nodes" %in% input$formatting) { | |||
gr <- select_nodes_by_id(gr, match(seeds@data$Node[seeds@data$Topic=="Selitykset"], gr$nodes_df$Oldid)) | |||
} | |||
gr <- deselect_nodes(gr, match(input$ignoreobj, gr$nodes_df$type)) | |||
if(verbose) cat("Selected nodes: ", gr$nodes_df$label[gr$nodes_df$id %in% gr$node_selection[[1]]]) | |||
return(gr) | |||
return( | |||
} | } | ||
objects.store( | objects.store(chooseGr) | ||
cat(" | cat("Ovariable chooseGr stored.\n") | ||
</rcode> | </rcode> | ||
Function insightJSON fetches a JSON file of an insight network through a REST API. Works on own computer only. | |||
<rcode name=" | <rcode name="insightJSON" label="Initiate function insightJSON (run on own computer)" embed=1> | ||
# This is code Op_en3861/ | # This is code Op_en3861/insightJSON on page [[Insight network]] | ||
#' | #' This function fetches an insight network data as JSON through REST api and makes a graph | ||
#' @param | #' @param URL URL for the insight network data | ||
#' @return a diagrammer graph object | |||
#' @return a | |||
insightJSON <- function(URL) { | |||
) { | |||
require(OpasnetUtils) | require(OpasnetUtils) | ||
require(DiagrammeR) | |||
require(jsonlite) | |||
objects.latest("Op_en3861", "formatted") # [[Insight network]] formatted | |||
tst <- fromJSON(url(URL)) | |||
nodes <- tst$data$nodes | |||
nodes <- data.frame( | |||
type = paste(nodes$indicator_level, nodes$type), | |||
Item = nodes$name, | |||
oldid = nodes$id, | |||
label = substr(nodes$name,1,30), | |||
tooltip = paste0(nodes$name,". ",nodes$id), | |||
URL = gsub("aplans.api","hnh",gsub("v1/","",nodes$url)), | |||
stringsAsFactors = FALSE | |||
) | |||
nodes$type <- gsub("NA ","",nodes$type) | |||
nodes <- merge(nodes, formatted, by.x="type", by.y="Resource") | |||
nodes$id <- 1:nrow(nodes) | |||
nodes <- nodes[!grepl("edge.",colnames(nodes))] | |||
colnames(nodes) <- gsub("node.","",colnames(nodes)) | |||
nodes <- nodes[c("id",setdiff(colnames(nodes),"id"))] | |||
edges <- tst$data$edges | |||
edges$from <- match(edges$from, nodes$oldid) | |||
edges$to <- match(edges$to, nodes$oldid) | |||
edges$oldid <- edges$id | |||
edges$rel <- gsub("_", " ", edges$effect_type) | |||
edges$label <- edges$rel | |||
edges$tooltip <- paste0(edges$rel, " (", edges$confidence_level,") ", edges$id) | |||
edges <- merge(edges, formatted, by.x="rel", by.y="Resource") | |||
edges$id <- 1:nrow(edges) | |||
edges <- edges[!grepl("node.",colnames(edges))] | |||
colnames(edges) <- gsub("edge.","",colnames(edges)) | |||
edges <- edges[c("from","to",setdiff(colnames(edges),c("from","to")))] | |||
gr <- create_graph(nodes, edges) | |||
return(gr) | |||
return( | |||
} | } | ||
objects.store( | #objects.store(insightJSON) # NOT STORED. | ||
cat("Function | #cat("Function insightJSON stored.\n") | ||
</rcode> | </rcode> | ||
==== | ==== Format tables ==== | ||
<rcode name="splizzeria" label="Initiate splizzeria (for developers only)" embed=1> | |||
# This is code Op_en3861/splizzeria on page [[Insight network]] | |||
library(OpasnetUtils) | |||
#' Split cells contents into vectors | |||
#' | |||
#' splizzeria function takes a data.frame and splits entries in cells of certain columns into separate rows. The idea is to make entries easier. | |||
#' | |||
#' @param df data.frame to be splitted | |||
#' @param cols names of columns that have the splittable contents | |||
#' @param split splitting character that separates individual entries in the cells | |||
#' @return data.frame with the same columns but (possibly) more rows than df. | |||
<rcode name=" | splizzeria <- function( | ||
# This is code Op_en3861/ | df, | ||
cols, | |||
split="," | |||
) { | |||
require(reshape2) | |||
for(i in cols) { | |||
d <- as.character(df[[i]]) | |||
d[d==""] <- NA # Because "" is incorrectly strsplitted | |||
d <- melt(strsplit(d, split=split), value.name=i) | |||
df$L1 <- 1:nrow(df) | |||
df <- merge(df[colnames(df)!=i], d) | |||
df[[i]] <- trimws(df[[i]]) | |||
df[is.na(df[[i]]),i] <- "" | |||
df$L1 <- NULL | |||
} | |||
return(df) | |||
} | |||
objects.store(splizzeria) | |||
cat("Function splizzeria stored.\n") | |||
</rcode> | |||
<rcode name="fillprev" label="Initiate function fillprev (for developers only)" embed=1> | |||
# This is code Op_en3861/fillprev on page [[Insight network]] | |||
library(OpasnetUtils) | library(OpasnetUtils) | ||
#' Filling empty cells | |||
#' | |||
#' fillprev fills empty cells in a data.frame by using content from the previous row. | |||
#' | |||
#' @param df data.frame to be filled | |||
#' @param cols vector of column names or positions to be filled. | |||
#' @return Returns a data.frame with the same shape as df. | |||
fillprev <- function(df, cols) { | |||
out <- df | |||
for(i in cols) { | |||
for(j in 2:nrow(out)) { | |||
if(out[j,i] %in% c("", NA)) out[j,i] <- out[j-1,i] | |||
} | |||
} | } | ||
) | return(out) | ||
} | |||
objects.store( | objects.store(fillprev) | ||
cat(" | cat("Function fillprev stored.\n") | ||
</rcode> | </rcode> | ||
==== Shiny server ==== | ==== Shiny server ==== | ||
<rcode name=" | <rcode name="ui" label="Initiate function ui (for developers only)" embed=1> | ||
# This is code Op_en3861/ | # This is code Op_en3861/ui on page [[Insight network]] | ||
library(OpasnetUtils) | library(OpasnetUtils) | ||
| Line 840: | Line 841: | ||
} | } | ||
objects.store(ui) | |||
cat("Function ui stored (makeUi is depreciated and not stored). Usage: shinyApp(ui, server, enableBookmarking = 'url')\n") | |||
objects.store(ui | |||
cat("Function ui stored (makeUi is | |||
</rcode> | </rcode> | ||
| Line 849: | Line 848: | ||
# This is code Op_en3861/server on page [[Insight diagram]]. | # This is code Op_en3861/server on page [[Insight diagram]]. | ||
library(OpasnetUtils) | library(OpasnetUtils) | ||
objects.latest("Op_en3861", code_name="chooseGr") # [[Insight network]] chooseGr | |||
#### Create shiny server at file server.R | #### Create shiny server at file server.R | ||
server <- function(input, output, session) { | server <- function(input, output, session) { | ||
output$plot1 <- renderGrViz({ | output$plot1 <- renderGrViz({ | ||
gr2 <- chooseGr(gr = gr, input = input, seeds = seeds) | |||
# gr <- deselect_nodes(gr, union( # This should find the INTERCEPT of from is selected AND to is selected and rel %in% ignorerel. | # gr <- deselect_nodes(gr, union( # This should find the INTERCEPT of from is selected AND to is selected and rel %in% ignorerel. | ||
# However, it is more complicated than that, because we may not want both from and to to disappear, only the one who is further away from core. | # However, it is more complicated than that, because we may not want both from and to to disappear, only the one who is further away from core. | ||
| Line 884: | Line 862: | ||
# tmp$edges_df$to[tmp$edges_df$rel %in% input$ignorerel] | # tmp$edges_df$to[tmp$edges_df$rel %in% input$ignorerel] | ||
# )) | # )) | ||
gr2$nodes_df$label <- gsub("(.{1,18})(\\s|$)", "\\1\n", gr2$nodes_df$label) # Cut labels to max 18 characters long on one line (except if a word is longer) | gr2$nodes_df$label <- gsub("(.{1,18})(\\s|$)", "\\1\n", gr2$nodes_df$label) # Cut labels to max 18 characters long on one line (except if a word is longer) | ||
# Alternative possibility is to use strwrap function from {base} or stri_wrap from stringi. | # Alternative possibility is to use strwrap function from {base} or stri_wrap from stringi. | ||
if("Hide node labels" %in% input$formatting) gr2$nodes_df$label <- "" | if("Hide node labels" %in% input$formatting) gr2$nodes_df$label <- "" | ||
if("Hide edge labels" %in% input$formatting) gr2$edges_df$label <- " " | if("Hide edge labels" %in% input$formatting) gr2$edges_df$label <- " " | ||
grViz(generate_dot(transform_to_subgraph_ws(gr2))) | grViz(generate_dot(transform_to_subgraph_ws(gr2))) | ||
}) | }) | ||
| Line 909: | Line 882: | ||
} | } | ||
objects.store(server) | objects.store(chooseGr, server) | ||
cat(" | cat("Functions chooseGr, server stored. Note! ChooseGr comes from its own code. Usage: shinyApp(ui, server, enableBookmarking = 'url')\n") | ||
</rcode> | </rcode> | ||
==== Scrape functions ==== | ==== Scrape functions ==== | ||
These functions | These functions were be placed in the OpasnetUtils package, which is [https://github.com/jtuomist/OpasnetUtils/blob/master/R/scrape.R maintained in Github]. To use the code, install a new version of the package by running R code | ||
devtools::install_github("jtuomist/OpasnetUtils") | |||
Codes Op_en3861/scrape.discussion, Op_en3861/scrape.functions, and Op_en3861/scrape.assessment on this page are outdated. | |||
scrape. | |||
==== Copy descriptions to ovariables ==== | ==== Copy descriptions to ovariables ==== | ||
| Line 1,475: | Line 1,046: | ||
== See also == | == See also == | ||
* [http://en.opasnet.org/en-opwiki/index.php?title=Insight_network&oldid=42630 Arhived version] 15.1.2019 with several functionalities that are now depreciated and removed. | |||
** T2b table [http://en.opasnet.org/w/Special:Opasnet_Base?id=op_en3861.table_types Table types] for different kinds of input tables. | |||
** Code for function grspec. This is no longer needed as a generic formatted data.frame is used for formatting of all resources. | |||
** Code for makeInsightGraph. This is replaced by makeGraph that has a better work flow. | |||
** Code for makeInsightTables. Insighttables are no longer produced as they are replaced by context-specific ovariables that are on their respective knowledge crystal pages. | |||
** Code for ovariable insightNetwork, which is an ovariable collecting all objects needed. Because of major updates, this is no longer useful. | |||
** Code server: function chooseGr was updated and moved to an own code. | |||
* [[Open policy practice]] | * [[Open policy practice]] | ||
Latest revision as of 20:13, 11 August 2021
| Moderator:Jouni (see all) |
|
|
| Upload data
|
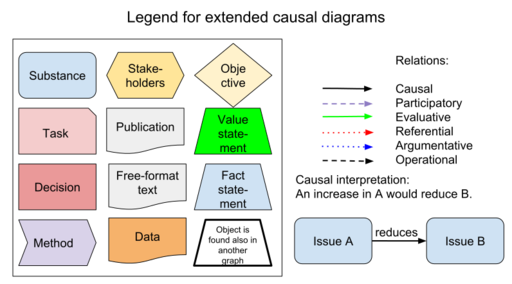
Insight networks are graphical representations of a particular situation, where the objects described are causally related to each other. In addition, the diagrams contain non-causal elements such as value judgements or inferences based on data. Insight networks utilise the ideas of directed acyclic graphs, but they have additional features.
Question
What notation is simple and flexible enough so that it can be used to represent all major issues related to a policy situation? It must be usable in both graphical and data formats.
Answer
- For examples of using insight networks, see op_fi:Ympäristöterveysindikaattori.
Rationale
Process
- Insight networks have been described in a scientific article manuscript From open assessment to shared understanding: practical experiences#Insight networks. Objects and their relations used in open policy practice are described on page Open policy ontology.
There is a need for methods facilitating the flow of information and understanding between science and policy. The principle is to describe a risk situation in a formal manner. Insight networks contain items along a causal pathway (or network) from e.g. abatement strategies to emissions to dispersion to exposure to effects. They have been designed to describe also non-causal connections such as non-causal reasoning, values, preferences, and arguments.
These diagrams use graph theory with vertices (or nodes) and arcs (or arrows). They are used to describe and define all the pieces needed for a description of the situation under scrutiny. Diagrams may be produced with any graphics software, providing that calculation functions are not required. If calculations are needed, we recommend the use of R software and OpasnetUtils package.
This is the process how data flows into insight diagrams:
- List of data tables of different insight diagrams is found from https://yhteistyotilat.fi/wiki08/x/1oGxAg. It has the following columns:
- Ilmio: Name of the phenomenon. This will become the name of a csv data file.
- Id: Identifier of the phenomenon. This will be used in Oldid of the items and relations.
- Tyyppi: Type of the table. In practice, it defines the columns that the data table has. Different types are listed on #Types of insight network tables.
- URL: Location of the data table. If the URL contains "google.com", it is assumed to be a google sheet. If the type (Tyyppi) is "keskustelu", it is assumed to be an Opasnet page with discussions. Otherwise, it is assumed to be a table on a web page that can be scraped with read_html() function.
- Taulu: If the data is a table on a web page, it is the number of the table on that page. If the data is a discussion, it is the number of discussion; missing value means that all discussions on that page are read.
- Alkurivi: In case of google sheets, it is the first row with actual data.
- Kuvaus: Description of the table, with possible links to relevant description page.
All data tables and discussions are listed, formatted and saved as csv files in a zip file called op_fi:File:Näkemysverkkojen tietotauluja.zip. From there, the data can be accessed from within Opasnet Rtools. (The code scraping web pages does not work in Opasnet, although it is stored there.) Little formatting is done here, mainly the column titles are standardised. But the number and type of columns is not changed.
In the next phase, each csv file is opened, interpreted, and defined as items and relations. This is done in code Op_fi5810/graphs on page op_fi:Ympäristöterveysindikaattori. All these are saved as a DiagrammeR graph, and each topic may be separately selected as a subgraph.
- 0094: . Are Tehtäväkokonaisuus, Osiotyyppi, JHS-luokka actually types of objects, or are they just indices. Yes, they should be indices and the objects relate to them with "has index". Correct table 4. --Jouni (talk) 20:46, 17 July 2018 (UTC) (type: ; paradigms: science: relevant attack)
- 0095: . Check the code about renderging graphs and creating a server function that also works on non-web environments on op_fi:Keskustelu:Näkemysverkko#Näkemysverkkoabstrakti vaikuttavuuden tutkimuksen päiville 4.-5.12.2018 --Jouni (talk) 20:46, 17 July 2018 (UTC) (type: ; paradigms: science: relevant attack)
Data
Graphical properties of objects and relations
| Obs | Property | Value | Parameter | Result | Description |
|---|---|---|---|---|---|
| 1 | default | default | node.shape | circle | Default values unless something else is specified |
| 2 | default | default | node.style | filled | |
| 3 | default | default | node.sides | 4 | |
| 4 | default | default | node.skew | 0 | |
| 5 | default | default | node.fillcolor | white | |
| 6 | default | default | node.fontsize | 11 | |
| 7 | default | default | node.height | 0.5 | |
| 8 | default | default | node.width | 0.5 | |
| 9 | default | default | node.color | brown | |
| 10 | default | default | node.penwidth | 2 | |
| 11 | default | default | node.fontcolor | black | |
| 12 | default | default | node.distortion | 0 | |
| 13 | default | default | edge.color | grey | |
| 14 | default | default | edge.fontsize | 10 | Not currently used |
| 15 | default | default | edge.fontcolor | grey | |
| 16 | default | default | edge.style | dotted | |
| 17 | default | default | edge.penwidth | 2 | |
| 18 | default | default | edge.arrowsize | 1 | Not currently used |
| 19 | type | unknown | node.fillcolor | yellow | This formatting is used if there are undefined objects |
| 20 | type | unknown | node.color | green | |
| 21 | type | substance | node.shape | circle | Substantive type object |
| 22 | type | substance | node.fillcolor | skyblue2 | Substantive type object |
| 23 | type | knowledge crystal | node.color | gold | Knowledge crystal type object (including ovariables and key ovariables) |
| 24 | type | option | node.color | palevioletred | Option for a decision |
| 25 | type | option | node.fillcolor | white | Option for a decision |
| 26 | type | index | node.shape | polygon | Index or other classifying determinant |
| 27 | type | index | node.sides | 4 | |
| 28 | type | index | node.skew | 0.5 | |
| 29 | type | index | node.fillcolor | purple1 | |
| 30 | type | index | node.height | 0.3 | |
| 31 | type | graph | node.shape | polygon | Index or other classifying determinant |
| 32 | type | graph | node.sides | 3 | |
| 33 | type | graph | node.fillcolor | pink | |
| 34 | type | assessment | node.shape | polygon | Assessment |
| 35 | type | assessment | node.sides | 8 | |
| 36 | type | assessment | node.fillcolor | purple1 | |
| 37 | type | stakeholder | node.shape | hexagon | Stakeholder type object |
| 38 | type | stakeholder | node.fillcolor | khaki1 | Stakeholder type object |
| 39 | type | stakeholder | node.width | 0.8 | Stakeholder type object |
| 40 | type | method | node.shape | polygon | Method type object |
| 41 | type | method | node.sides | 6 | Method type object |
| 42 | type | method | node.fillcolor | purple1 | Method type object |
| 43 | type | process | node.shape | pentagon | Process type object |
| 44 | type | process | node.fillcolor | purple1 | Process type object |
| 45 | type | action | node.fillcolor | #009246 | Process type object, dark green (0,146,70) |
| 46 | type | action | node.shape | rectangle | Decision type object |
| 47 | type | task 1 | node.color | brown | Illustration of the responsible organisation of the task |
| 48 | type | task 2 | node.color | yellow | Illustration of the responsible organisation of the task |
| 49 | type | task 3 | node.color | blue | Illustration of the responsible organisation of the task |
| 50 | type | task 4 | node.color | green | Illustration of the responsible organisation of the task |
| 51 | type | task 5 | node.color | red | Illustration of the responsible organisation of the task |
| 52 | type | decision | node.fillcolor | red | Decision type object |
| 53 | type | data | node.shape | rectangle | Data type object |
| 54 | type | data | node.fillcolor | gold | Data type object |
| 55 | type | objective | node.shape | diamond | Objective type object |
| 56 | type | objective | node.fillcolor | yellow | Objective type object |
| 57 | type | objective | node.width | 0.8 | Objective type object |
| 58 | type | publication | node.fillcolor | gray | Publication type object |
| 59 | type | statement | node.shape | polygon | Argument type object |
| 60 | type | statement | node.sides | 4 | Argument type object |
| 61 | type | statement | node.width | 0.8 | Argument type object |
| 62 | type | statement | node.distortion | -0.5 | Argument type object |
| 63 | type | true statement | node.fillcolor | gold | Argument type object |
| 64 | type | false statement | node.fillcolor | gray | Argument type object |
| 65 | type | fact opening statement | node.fillcolor | lightskyblue1 | Argument type object. Discussion start |
| 66 | type | value opening statement | node.fillcolor | palegreen1 | Argument type object |
| 67 | type | fact closing statement | node.fillcolor | skyblue | Argument type object. Discussion end |
| 68 | type | value closing statement | node.fillcolor | springgreen | Argument type object. |
| 69 | type | fact discussion | node.fillcolor | skyblue | Argument type object. Not neede? |
| 70 | type | value discussion | node.fillcolor | springgreen | Value judgement type object. Not needed? |
| 71 | type | risk factor | node.color | pink | Additional information about object class |
| 72 | type | indicator | node.color | brown | Additional information about object class |
| 73 | type | indicator | node.fillcolor | gold | Additional information about object class |
| 74 | type | operational indicator | node.fillcolor | #00d7a7 | Additional information about object class light green (0,215,167) |
| 75 | type | tactical indicator | node.fillcolor | #9fc9eb | Additional information about object class light blue (159,201,235) |
| 76 | type | strategic indicator | node.fillcolor | #0072c6 | Additional information about object class dark blue (0,114,198) |
| 77 | type | strategic indicator | node.shape | diamond | Additional information about object class |
| 78 | type | arviointikriteeri | node.color | orange | Not quite clear what criteria objects are: indicators or value statements, or something else |
| 79 | type | task | node.color | green | Additional information about object class |
| 80 | type | data | node.color | orange | Additional information about object class |
| 81 | type | health organisation | node.color | yellow | Additional information about object class |
| 82 | Relation | causal link | edge.color | black | Causal link |
| 83 | Relation | causal link | edge.style | solid | Causal link |
| 84 | Relation | positive causal link | edge.fontcolor | #009246 | Causal link, dark green (0,146,70) |
| 85 | Relation | increases | edge.fontcolor | #009246 | Causal link, dark green (0,146,70) |
| 86 | Relation | negative causal link | edge.fontcolor | #bd2719 | Causal link, red (189,39,25) |
| 87 | Relation | decreases | edge.fontcolor | #bd2719 | Causal link, red (189,39,25) |
| 88 | Relation | part_of | edge.fontcolor | gray | Part of (set theory link) |
| 89 | Relation | participatory link | edge.color | purple | Participatory link |
| 90 | Relation | participatory link | edge.style | dashed | Participatory link |
| 91 | Relation | operational link | edge.color | black | Operational link |
| 92 | Relation | operational link | edge.style | dashed | Operational link |
| 93 | Relation | evaluative link | edge.color | green | Evaluative link |
| 94 | Relation | relevant attack | edge.color | red | Attacking argument |
| 95 | Relation | relevant defense | edge.color | green | Defending argument |
| 96 | Relation | relevant comment | edge.color | blue | Commenting argument |
| 97 | Relation | irrelevant argument | edge.color | gray | Invalid argument |
| 98 | Relation | argumentative link | edge.style | dotted | Argumentative link |
| 99 | Relation | argumentative link | edge.penwidth | 4 | Argumentative link |
| 100 | Relation | referential link | edge.color | red | Referential link |
| 101 | Relation | referential link | edge.style | dashed | Referential link |
Calculations
Insight network 2.0
An updated version should improve the
- a) context sensitivity (referring to primarily to objects within own context but secondarily to those from another context),
- b) making graphs by default from a single context rather than a full list of contexts from a meta table,
- c) compatibility with cytoscape.js,
- d) merging ready-made graphs meaningfully,
- e) have a reasonable intermediate object format that contains all data needed, such as
- tables for nodes and edges, compatible with Diagrammer, Cytoscape.js, AND Gephi.
- metadata for display, such as seeds, steps, object types to ignore, whether to show labels etc. Or should these just be implemented on the graph?
What should be done?
- Fetch the data table by scrape or other function and with data about URL, table, and initial row.
- Use splizzeria and fillprev if needed.
- Interpret columns based on a vector of column numbers (with possibly 1+2 notation to paste columns) to create the standard columns. If this is done in an ovariable formula, there is no need for a specific function.
- Context
- Item
- type
- label
- rel
- Object
- Description
- Reldescription
- URL
- Result (dummy, always 0)
- Create missing node rows from objects. Do NOT assume context.
- Create URL from permanent resource location trunk and the identifier (where does the identifier come from?)
- Item ja label laitetaan pötköön ja haetaan mätsi. Tulos onrow-pötköstä.
- Create an ovariable from the table.
- Add meta to the ovariable with formatting data.
- insightGraph:
- seed
- removenodes
- formatting (character vector with possible entries: Hide node labels, Hide edge labels, Show legend nodes, Remove branches only)
- ignoreobj
- steps
- insightGraph:
- (NOT NEEDED? Create Oldid if does not exist from context and numbering)
- If a relation is presented as item, the formatting is applied to the ring.
Combine graph objects
- Find items without context. Match them with items with the same Item (label) that do have a type.
Tuplarelaatiot, voidaanko kategorisesti poistaa?
Out <- rep(NA, length(find))
For(x in cond,)
For(i in 1:length(find)
Tmp<-id[context==contextfind(i))])[Match(find(i), df$cond(x)(df$context==contextfind(i))] pitää etsiä id alkuperäisestä taulukosta heti muuten ei toimi
Out<- ifelse(isna(out). Tmp,out)
))
Sitten sama ioman contekstirajoitusta.
Insight network 1.0
There are three different identifiers for a subject item.
- Oldid: a technical identifier typically of format context.number, where number is a sequential number within a context.
- Item: the actual name of the item, detailed enough to give a good understanding of its meaning.
- label: a short name shown on insight networks. Does not exmplain everything, just enough to distinguish it from other items.
If Oldid is not given, it is created from the context and a number. If label is not given in data, it is truncated from Item.
Object item has one column Object that may contain any of these. The priority is Item > label > Oldid > Object. The last option means that it is assumed that Object refers to a new item that is not mentioned in the Item column.
An insight network is produced in this order (last object mentioned first).
- gr: a diagrammer graph with all data and formatting for an insight network. Produced by makeInsightGraph.
- makeInsightGraph
Making insight graphs
Function insightJSON fetches a JSON file of an insight network through a REST API. Works on own computer only.
Format tables
Shiny server
Scrape functions
These functions were be placed in the OpasnetUtils package, which is maintained in Github. To use the code, install a new version of the package by running R code
devtools::install_github("jtuomist/OpasnetUtils")
Codes Op_en3861/scrape.discussion, Op_en3861/scrape.functions, and Op_en3861/scrape.assessment on this page are outdated.
Copy descriptions to ovariables
The function assessmentDescriptions scans through an assessment ovarible that has all relevant assessment objects as parents. Dependencies slot may also have additional information, such as the following.
- Name: name of parent (obligatory)
- Ident: Opasnet page identifier and code name where the parent object can be loaded (e.g. Op_en7748/hia). Note: This is typically the code for the whole assessment, not the individual codes for the objects.
- Token: Token for the model run where the parent object can be loaded (e.g. xxNsLw5hWdM6xyYp)
- Description: A short description about what the object is. This is typically shown when cursor hovers over the object on an online insight diagram.
- Page: Opasnet page identifier for the object's knowledge crystal page, which contains the research question, answer, and description of the object, together with discussion, if any. Typically this is empty for ovariables, because this information can be found from ovariable@meta slot and there is no need to duplicate it here.
- Child: An object to which this object links. This is typically needed for objects such as graphs and data.frames that do not contain this information in their own structure, unlike ovariables. The direction of a relation is away from this object because then this object is the subject in triple sentences and can be given other parameters as well in other columns. A typical sentence is "graph describes ovariable", but for illustrative purposes this is inversed on insight networks so that the arrow points from an ovariable to a graph ("ovariable is described by graph").
- Other columns are allowed.
Old notation
⇤--#: . Look at the table below together with Open policy ontology and merge. Decide which things should be on this page and which should be on the other. --Jouni (talk) 06:55, 24 April 2018 (UTC) (type: truth; paradigms: science: attack)
Previous notations
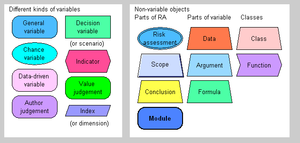
Insight networks have previously been called pyrkilo diagrams, extended causal diagrams, and factor-effect-value networks. These names are no longer in active use. An archived version of the notation can be found from an earlier version of this page.
See also
- Arhived version 15.1.2019 with several functionalities that are now depreciated and removed.
- T2b table Table types for different kinds of input tables.
- Code for function grspec. This is no longer needed as a generic formatted data.frame is used for formatting of all resources.
- Code for makeInsightGraph. This is replaced by makeGraph that has a better work flow.
- Code for makeInsightTables. Insighttables are no longer produced as they are replaced by context-specific ovariables that are on their respective knowledge crystal pages.
- Code for ovariable insightNetwork, which is an ovariable collecting all objects needed. Because of major updates, this is no longer useful.
- Code server: function chooseGr was updated and moved to an own code.
- Open policy practice
- Open policy ontology
- From open assessment to shared understanding: practical experiences
- Category:Causal diagram