Assessment of the health impacts of H1N1 vaccination: Difference between revisions
m (→R code: modifications to notes in code) |
mNo edit summary |
||
| (19 intermediate revisions by 4 users not shown) | |||
| Line 1: | Line 1: | ||
[[Category:DARM exercise]] | [[Category:DARM exercise]] | ||
{{assessment|moderator=Teemu R}} | [[Category:Online model]] | ||
[[Category:THL publications 2011]] | |||
{{assessment|moderator=Teemu R | |||
| reference = {{publication | |||
| authors = Teemu Rintala, Jouni T. Tuomisto, Mikko V. Pohjola, Kati Iso-Markku, Virpi Kollanus, Marko Tainio et al. | |||
| page = Assessment of the health impacts of H1N1 vaccination | |||
| explanation = Comparison of swine flu vaccination strategies in Finland | |||
| publishingyear = 2011 | |||
| urn = | |||
| elsewhere = | |||
}} | |||
}} | |||
[[op_fi:Sikainfluenssarokotteen terveyshaitat]] | [[op_fi:Sikainfluenssarokotteen terveyshaitat]] | ||
<big> This assessment was completed in 2011, it is no longer actively updated. </big> | |||
{{summary box | {{summary box | ||
|question = What was the overall health impact of the H1N1 (swine flu) vaccination in Finland in 2009-2010? Given current knowledge, which was the better decision between vaccinating as happened versus vaccinating no-one versus not vaccinating the population aged 5-19? | |question = What was the overall health impact of the H1N1 (swine flu) vaccination in Finland in 2009-2010? Given current knowledge, which was the better decision between vaccinating as happened versus vaccinating no-one versus not vaccinating the population aged 5-19? | ||
|answer = Given current knowledge, the decision to vaccinate the whole population was the best decision even when narcolepsy is included in the assessment. Results of the [[Value of information]] analysis suggest that further knowledge about the uncertain variables considered very likely would not have changed the decision.}} | |answer = Given current (2011) knowledge, the decision to vaccinate the whole population was the best decision even when narcolepsy is included in the assessment. Results of the [[Value of information]] analysis suggest that further knowledge about the uncertain variables considered very likely would not have changed the decision.}} | ||
This assessment is about the total health effects the 2009 swine flu pandemic. It utilizes data from the Infectious disease registry (TTR) maintained by THL, the National narcolepsy task force report from 31.1.2011 and WHO. The current model is a simplification with no time dimension. The assessment has been evaluated. Evaluation results are displayed on the [[Talk:Assessment of the health impacts of H1N1 vaccination#Evaluation of the H1N1 assessment|discussion page]] | |||
This assessment is about the total health effects the 2009 swine flu pandemic. It utilizes data from the Infectious disease registry (TTR) maintained by THL, the National narcolepsy task force report from 31.1.2011 and WHO. The current model is a simplification with no time dimension. | |||
==Scope== | ==Scope== | ||
| Line 65: | Line 76: | ||
*[[Disability weights]] | *[[Disability weights]] | ||
**DALY weight of narcolepsy equals roughly that of epilepsy (0.065) | **DALY weight of narcolepsy equals roughly that of epilepsy (0.065) | ||
**DALY weight of | **DALY weight of swine flu for the duration of the disease ~ 0.5 (~ 0.007 DALYs per case when adjusted for duration) | ||
*Life expectancy by age groups in Finland<ref>[http://www.who.int/healthinfo/statistics/mortality_life_tables/en/ WHO life table estimates for 2008, Finland]</ref> | *Life expectancy by age groups in Finland<ref>[http://www.who.int/healthinfo/statistics/mortality_life_tables/en/ WHO life table estimates for 2008, Finland]</ref> | ||
*Probability of catching swine flu given subject is not immune | *Probability of catching swine flu given subject is not immune | ||
| Line 77: | Line 88: | ||
*Fraction of population belonging to a risk group | *Fraction of population belonging to a risk group | ||
**Arbitrary values; trying to account for kids of age <1 and old folks with heart conditions etc. | **Arbitrary values; trying to account for kids of age <1 and old folks with heart conditions etc. | ||
* | *Duration of swine flu | ||
**Assumed to be flat 5 days (mildly incapacitated for this duration) | **Assumed to be flat 5 days (mildly incapacitated for this duration) | ||
*[[Narcolepsy in Finland]] | *[[Narcolepsy in Finland]] | ||
| Line 92: | Line 103: | ||
**Uncertainties of ERF of vaccine on narcolepsy, fraction of all cases represented by lab confirmed cases and probability of catching swine flu are implemented. | **Uncertainties of ERF of vaccine on narcolepsy, fraction of all cases represented by lab confirmed cases and probability of catching swine flu are implemented. | ||
<rcode graphics="1"> | <rcode graphics="1" variables="name:narcweight|description:Disability weight for narcolepsy|default:0.065| | ||
name:n|description:How many iterations do you want to run? (Full model is 1000 but it is slow)|default:200"> | |||
# Model; original data inputs are disability weights (isfw, inarcw), population (ipop/dpop), (atm) ERF of vaccine on narcolepsy (NERF), | # Model; original data inputs are disability weights (isfw, inarcw), population (ipop/dpop), (atm) ERF of vaccine on narcolepsy (NERF), | ||
# vaccination coverage (ivac_cov), base immunity in the population (ibimm) and observed number of sf cases (dsf) | # vaccination coverage (ivac_cov), base immunity in the population (ibimm) and observed number of sf cases (dsf) | ||
| Line 105: | Line 117: | ||
# iElrgc is the expected life correction for the risk group (people with heart conditions etc are likely to die earlier anyway) | # iElrgc is the expected life correction for the risk group (people with heart conditions etc are likely to die earlier anyway) | ||
outcome <- function(inarc = narc(), isf = sf(), inarcw = data.frame(Result= | outcome <- function(inarc = narc(), isf = sf(), inarcw = data.frame(Result=narcweight),#op_baseGetData("opasnet_base", "Op_en2307"), | ||
isfw = data.frame(Result=0.5), iEl = data.frame(Age=c("0-4 ","5-9","10-14","15-19","20-24","25-29","30-34","35-39","40-44","45-49", | isfw = data.frame(Result=0.5), iEl = data.frame(Age=c("0-4","5-9","10-14","15-19","20-24","25-29","30-34","35-39","40-44","45-49", | ||
"50-54","55-59","60-64","65-69","70-74","75-79","80+","All"), Result = c(79,75.1,70.1,65.1,60.3,55.5,50.7,45.9,41.1,36.5,31.9,27.6, | "50-54","55-59","60-64","65-69","70-74","75-79","80+","All"), Result = c(79,75.1,70.1,65.1,60.3,55.5,50.7,45.9,41.1,36.5,31.9,27.6, | ||
23.5,19.5,15.7,12.1,9,50)), ilsf = data.frame(Result=5/365), isfpd = sfpd(), ifrg = data.frame(Age = c("0-4 ","5-9","10-14","15-19", | 23.5,19.5,15.7,12.1,9,50)), ilsf = data.frame(Result=5/365), isfpd = sfpd(), ifrg = data.frame(Age = c("0-4","5-9","10-14","15-19", | ||
"20-24","25-29","30-34","35-39","40-44","45-49","50-54","55-59","60-64","65-69","70-74","75-79","80+"), Result = c(0.1, rep(0.001, | "20-24","25-29","30-34","35-39","40-44","45-49","50-54","55-59","60-64","65-69","70-74","75-79","80+"), Result = c(0.1, rep(0.001, | ||
11), rep(0.5,5))), iElrgc = data.frame(Age = c("0-4 ","5-9","10-14","15-19","20-24","25-29","30-34","35-39","40-44","45-49","50-54", | 11), rep(0.5,5))), iElrgc = data.frame(Age = c("0-4","5-9","10-14","15-19","20-24","25-29","30-34","35-39","40-44","45-49","50-54", | ||
"55-59","60-64","65-69","70-74","75-79","80+"), Result = c(rep(1,12), rep(0.5,5))), ...) { | "55-59","60-64","65-69","70-74","75-79","80+"), Result = c(rep(1,12), rep(0.5,5))), ...) { | ||
| Line 157: | Line 169: | ||
# Function for calculating narcolepsy cases, depends on erf of vac on narc, pop, vac_cov and background incidence (fraction) | # Function for calculating narcolepsy cases, depends on erf of vac on narc, pop, vac_cov and background incidence (fraction) | ||
narc <- function(iNERF = op_baseGetData("opasnet_base", "Op_en4923") | narc <- function(iNERF = op_baseGetData("opasnet_base", "Op_en4923"), ipop = op_baseGetData("opasnet_base", "Op_en2949"), | ||
ivac_cov = op_baseGetData("opasnet_base", "Op_en4925") | ivac_cov = op_baseGetData("opasnet_base", "Op_en4925"), ibgi = data.frame(Result=1.4e-5), ...) { | ||
iNERF <- iNERF[, !colnames(iNERF)%in%c("id", "Result.Text")] | |||
ipop <- ipop[, !colnames(ipop)%in%c("id", "obs", "Result.Text")] | |||
ivac_cov <- ivac_cov[, !colnames(ivac_cov)%in%c("id", "obs", "Result.Text")] | |||
names(iNERF)[names(iNERF)=="Result"] <- "iNERF" | names(iNERF)[names(iNERF)=="Result"] <- "iNERF" | ||
names(ipop)[names(ipop)=="Result"] <- "ipop" | names(ipop)[names(ipop)=="Result"] <- "ipop" | ||
| Line 175: | Line 190: | ||
# Function for calculating swine flu cases | # Function for calculating swine flu cases | ||
# ilcf is the fraction of actual swine flu cases represented by the lab confirmed cases | # ilcf is the fraction of actual swine flu cases represented by the lab confirmed cases | ||
# (scrapped)assumed binomially distributed, if uncertainty of probability of getting swine flu is not | # (scrapped)assumed binomially distributed, if uncertainty of probability of getting swine flu is not | ||
# present, the point estimate will be used as such and n samples sampled from the same distribution; | # present, the point estimate will be used as such and n samples sampled from the same distribution; | ||
# however if the probability has an uncertainty, only 1 sample from each distribution is sampled where | # however if the probability has an uncertainty, only 1 sample from each distribution is sampled where | ||
# samples of the probability will be used as inputs; disabled sampling here since it would make different scenarios incomparable | # samples of the probability will be used as inputs;(/scrapped) disabled sampling here since it would make different scenarios incomparable | ||
sf <- function(isfp = sfp(), ipop = op_baseGetData("opasnet_base", "Op_en2949") | sf <- function(isfp = sfp(), ipop = op_baseGetData("opasnet_base", "Op_en2949"), iimm = imm(), n = 1000, | ||
ilcf = | ilcf = data.frame(Result = 0.2), ...) { | ||
ipop <- ipop[, !colnames(ipop)%in%c("id", "obs", "Result.Text")] | |||
names(isfp)[names(isfp)=="Result"] <- "isfp" | names(isfp)[names(isfp)=="Result"] <- "isfp" | ||
names(ipop)[names(ipop)=="Result"] <- "ipop" | names(ipop)[names(ipop)=="Result"] <- "ipop" | ||
| Line 206: | Line 222: | ||
# dsf, dpop and dimm are the actual data we have | # dsf, dpop and dimm are the actual data we have | ||
# uncertainty: number of cases is binomially distributed, number of trials (n) is the number of people who were not immune to swine-flu | # uncertainty: number of cases is binomially distributed, number of trials (n) is the number of people who were not immune to swine-flu | ||
# outcome | # outcome x is known, p is unknown. P can be easily calculated from x = p*n, when we assume that x is the expected value of the | ||
# distribution. But we're interested in the uncertainty of the estimate... When n is suitably large a poisson distribution becomes | # distribution. But we're interested in the uncertainty of the estimate... When n is suitably large a poisson distribution becomes | ||
# an excellent approximation of the binomial distribution. So we can fit the poisson distribution using an R function (I didn't find | # an excellent approximation of the binomial distribution. So we can fit the poisson distribution using an R function (I didn't find | ||
| Line 212: | Line 228: | ||
# rubbish because we only have a single observation, sd of the mean estimate of the poisson distribution appears to be (n*p)^0.5, so the | # rubbish because we only have a single observation, sd of the mean estimate of the poisson distribution appears to be (n*p)^0.5, so the | ||
# variance of the parameter is n * p, which divided by n gives p ... so the expected value is p and the variance is p, which causes problems | # variance of the parameter is n * p, which divided by n gives p ... so the expected value is p and the variance is p, which causes problems | ||
# with the beta distribution (shape parameters become negative), | # with the beta distribution (shape parameters become negative), so I've used standard error^4 as the variance... | ||
# Somebody with real knowledge about these things should check this sometime, I'm just playing around with some numbers to get more | |||
# uncertainty in the model. | |||
sfp <- function(dsf = op_baseGetData("opasnet_base", "Op_en4933") | sfp <- function(dsf = op_baseGetData("opasnet_base", "Op_en4933"), dpop = op_baseGetData("opasnet_base", "Op_en2949"), #series_id = 970), | ||
dimm = imm(), n = 1000, ...) { | |||
dsf <- dsf[, !colnames(dsf)%in%c("id", "obs", "Result.Text")] | |||
dpop <- dpop[, !colnames(dpop)%in%c("id", "obs", "Result.Text")] | |||
names(dsf)[names(dsf)=="Result"] <- "dsf" | names(dsf)[names(dsf)=="Result"] <- "dsf" | ||
names(dpop)[names(dpop)=="Result"] <- "dpop" | names(dpop)[names(dpop)=="Result"] <- "dpop" | ||
| Line 237: | Line 257: | ||
} | } | ||
# Function for calculating probability of mortality after catching swine flu given that | # Function for calculating probability of mortality after catching swine flu given that the subject belongs to a risk group | ||
# children of age < 1 and adults of 60+ afflicted by heart conditions etc are considered to belong to a risk group | # children of age < 1 and adults of 60+ afflicted by heart conditions etc are considered to belong to a risk group | ||
# ifrg is the fraction belonging to the risk group in a given age group | # ifrg is the fraction belonging to the risk group in a given age group | ||
# p(death|sf&rg)) assumed constant in all age groups | # p(death|sf&rg)) assumed constant in all age groups | ||
# dsf is the data we have on the number of swine flu cases, dmsf is the data on mortality caused by swine flu, dfrg is the data on | |||
# the fraction of population belonging to risk groups (arbitrary default values included) | |||
sfpd <- function(dsf = op_baseGetData("opasnet_base", "Op_en4933") | sfpd <- function(dsf = op_baseGetData("opasnet_base", "Op_en4933"), dmsf = data.frame(Result=44), | ||
dfrg = data.frame(Age = c("0-4 ","5-9","10-14","15-19","20-24","25-29","30-34","35-39","40-44","45-49", | dfrg = data.frame(Age = c("0-4 ","5-9","10-14","15-19","20-24","25-29","30-34","35-39","40-44","45-49", | ||
"50-54","55-59","60-64","65-69","70-74","75-79","80+"), Result = c(0.1, rep(0.001,11), rep(0.5,5))), ilcf = tlcf, ...) { | "50-54","55-59","60-64","65-69","70-74","75-79","80+"), Result = c(0.1, rep(0.001,11), rep(0.5,5))), ilcf = tlcf, ...) { | ||
dsf <- dsf[, !colnames(dsf)%in%c("id", "obs", "Result.Text")] | |||
names(dsf)[names(dsf)=="Result"] <- "dsf" | names(dsf)[names(dsf)=="Result"] <- "dsf" | ||
names(dmsf)[names(dmsf)=="Result"] <- "dmsf" | names(dmsf)[names(dmsf)=="Result"] <- "dmsf" | ||
| Line 262: | Line 285: | ||
# Function for calculating total immunity to swine flu in the whole population | # Function for calculating total immunity to swine flu in the whole population | ||
# P(imm) = P(vac or base_imm) | # P(imm) = P(vac or base_imm) | ||
# ibimm is the input background immunity | |||
# ivac_cov is the input vaccination coverage | |||
imm <- function(ibimm = op_baseGetData("opasnet_base", "Op_en4943", series_id = 994) | imm <- function(ibimm = op_baseGetData("opasnet_base", "Op_en4943"), #series_id = 994), | ||
ivac_cov = op_baseGetData("opasnet_base", "Op_en4925")[, | ivac_cov = op_baseGetData("opasnet_base", "Op_en4925"), ...) { | ||
ibimm <- ibimm[, !colnames(ibimm)%in%c("id", "obs", "Result.Text")] | |||
ivac_cov <- ivac_cov[, !colnames(ivac_cov)%in%c("id", "obs", "Result.Text")] | |||
names(ibimm)[names(ibimm)=="Result"] <- "ibimm" | names(ibimm)[names(ibimm)=="Result"] <- "ibimm" | ||
names(ivac_cov)[names(ivac_cov)=="Result"] <- "ivac_cov" | names(ivac_cov)[names(ivac_cov)=="Result"] <- "ivac_cov" | ||
| Line 273: | Line 300: | ||
} | } | ||
# Calculating the outcome with different scenarios | ######################################################################3 | ||
## Calculating the outcome with different scenarios | |||
# Libraries loaded and some basic functions defined | |||
paralpha <- function(imean, ivar) imean * (imean * (1 - imean) / ivar - 1) | paralpha <- function(imean, ivar) imean * (imean * (1 - imean) / ivar - 1) # beta distribution parameters | ||
parbeta <- function(imean, ivar) (1 - imean) * (imean * (1 - imean) / ivar - 1) | parbeta <- function(imean, ivar) (1 - imean) * (imean * (1 - imean) / ivar - 1) | ||
library( | library(OpasnetUtils) | ||
library(MASS) | library(MASS) | ||
library(ggplot2) | library(ggplot2) | ||
library(xtable) | |||
# Common variable generation | # Common variable generation | ||
n <- 1000 | #n <- 1000 | ||
tlcf <- data.frame(obs = 1:n, Result=rbeta(n, paralpha(0.2, 0.1^2), parbeta(0.2, 0.1^2))) | tlcf <- data.frame(obs = 1:n, Result=rbeta(n, paralpha(0.2, 0.1^2), parbeta(0.2, 0.1^2))) | ||
#ggplot(tlcf, aes(x=Result)) + geom_density() | #ggplot(tlcf, aes(x=Result)) + geom_density() | ||
| Line 295: | Line 326: | ||
# Default, model based on current knowledge | # Default, model based on current knowledge | ||
test <- op_baseGetData("opasnet_base", "Op_en4926") # Old run | |||
test <- test[, !colnames(test)%in%c("id", "Result.Text")] | |||
temp2 <- as.data.frame(as.table(apply(tapply(test$Result, test[,c("Age","Scenario","Outcome")], mean), c(2,3), sum))) | temp2 <- as.data.frame(as.table(apply(tapply(test$Result, test[,c("Age","Scenario","Outcome")], mean), c(2,3), sum))) | ||
ggplot(temp2, aes(x = | #ggplot(temp2, aes(x = Scenario, weight = Freq, fill = Outcome)) + geom_bar(position = "stack") # position = "dodge" | ||
#head(temp2) | |||
# For further analysis we need to use our recently generated lab confirmed fraction, this should be uploaded somewhere | |||
temp <- op_baseGetData("opasnet_base", "Op_en4925")[, | temp <- op_baseGetData("opasnet_base", "Op_en4925") # New run. Op_en4925 = Vaccination coverage in Finland | ||
tvac_cov <- data.frame(temp, Scenario = " | temp <- temp[, !colnames(temp)%in%c("id", "obs", "Result.Text")] | ||
tvac_cov <- rbind(tvac_cov, data.frame(Age=tvac_cov[,"Age"], Result = 0, Scenario = " | tvac_cov <- data.frame(temp, Scenario = "Vaccinate all") | ||
tvac_cov <- rbind(tvac_cov, data.frame(Age=tvac_cov[,"Age"], Result = 0, Scenario = "Vaccinate nobody")) | |||
temp[2:4,2] <- 0 | temp[2:4,2] <- 0 | ||
tvac_cov <- rbind(tvac_cov, data.frame(temp, Scenario = "5- | tvac_cov <- rbind(tvac_cov, data.frame(temp, Scenario = "Vaccinate all but 5-19 a")) | ||
test <- outcome(inarc = narc(ivac_cov = tvac_cov), sf(isfp = tsfp, iimm = imm(ivac_cov = tvac_cov))) | test <- outcome(inarc = narc(ivac_cov = tvac_cov), isf = sf(isfp = tsfp, iimm = imm(ivac_cov = tvac_cov), ilcf = tlcf)) | ||
test[test$Outcome=="onarc", "Outcome"] <- "Narcolepsy" | |||
test[test$Outcome=="osf", "Outcome"] <- "Swine flu morbidity" | |||
test[test$Outcome=="osfm", "Outcome"] <- "Swine flu mortality" | |||
temp <- as.data.frame(as.table(tapply(test$Result, test[,c("obs","Scenario")], sum))) | temp <- as.data.frame(as.table(tapply(test$Result, test[,c("obs","Scenario")], sum))) | ||
ggplot(temp, aes(x = Freq, fill = Scenario)) + geom_density(alpha = 0.2) | ggplot(temp, aes(x = Freq, fill = Scenario)) + geom_density(alpha = 0.2) | ||
tapply(temp$Freq, temp[,c("Scenario")], mean) | temp4 <- as.data.frame(as.table(tapply(temp$Freq, temp[,c("Scenario")], mean))) | ||
colnames(temp4) <- c("Decision option", "DALYs") | |||
print(xtable(temp4), type = 'html') | |||
ggplot(test, aes(x = Scenario, weight = Result, fill = Outcome)) + geom_bar(position = "stack") # position = "dodge" | |||
#op_baseWrite("opasnet_base", test) | #op_baseWrite("opasnet_base", test) | ||
| Line 328: | Line 357: | ||
#test <- op_baseGetData("opasnet_base", "Op_en4926") | #test <- op_baseGetData("opasnet_base", "Op_en4926") | ||
temp2 <- as.data.frame(as.table(tapply(test$Result, test[,(colnames(test)%in%c("Result","Outcome"))==FALSE], sum))) | temp2 <- as.data.frame(as.table(tapply(test$Result, test[,(colnames(test)%in%c("Result","Outcome"))==FALSE], sum))) | ||
tNERF <- op_baseGetData("opasnet_base", "Op_en4923")[, | tNERF <- op_baseGetData("opasnet_base", "Op_en4923") | ||
tNERF <- tNERF[, !colnames(tNERF)%in%c("id", "Result.Text")] | |||
colnames(tNERF)[colnames(tNERF)=="Result"] <- "tNERF" | colnames(tNERF)[colnames(tNERF)=="Result"] <- "tNERF" | ||
colnames(tlcf)[colnames(tlcf)=="Result"] <- "tlcf"#tlcf from above | colnames(tlcf)[colnames(tlcf)=="Result"] <- "tlcf"#tlcf from above | ||
| Line 357: | Line 387: | ||
## Correlation | ## Correlation | ||
cor(temp2$Freq, temp2[,c("tNERF", "tlcf", "tsfp")], method = "spearman") | correlations <- as.data.frame(as.table(cor(temp2$Freq, temp2[,c("tNERF", "tlcf", "tsfp")], method = "spearman")))[, 2:3] | ||
colnames(correlations) <- c("Variable", "Spearman correlation vs. outcome") | |||
correlations$Variable <- c("Narcolepsy ERF", "Fraction of lab-confirmed cases", "P(swine flu|non-immune)") | |||
print(xtable(correlations), type = 'html') | |||
## EVPI | ## EVPI | ||
mean(tapply(temp$Freq, temp$obs, min)) - min(tapply(temp$Freq, temp$Scenario, mean)) # not considering by age group | EVPI <- mean(tapply(temp$Freq, temp$obs, min)) - min(tapply(temp$Freq, temp$Scenario, mean)) # not considering by age group | ||
mean(apply(tapply(temp2$Freq, temp2[,c("obs","Age")], min), 1, sum)) - sum(apply(tapply(temp2$Freq, temp2[,c("Scenario","Age")], mean), 2, min)) | |||
EVPI.agegroup <- mean(apply(tapply(temp2$Freq, temp2[,c("obs","Age")], min), 1, sum)) - sum(apply(tapply(temp2$Freq, temp2[,c("Scenario","Age")], mean), 2, min)) # Not used in the table because I am not quite sure what this means | |||
## EVXPI for NERF, lcf and sfp | ## EVXPI for NERF, lcf and sfp | ||
mean(apply(tapply(temp2$EogNERF, temp2[,(colnames(temp2)%in%c("Freq","Scenario","tNERF","tlcf","tsfp","NERFbin","lcfbin","sfpbin", | EVXPI.NERF <- mean(apply(tapply(temp2$EogNERF, temp2[,(colnames(temp2)%in%c("Freq","Scenario","tNERF","tlcf","tsfp","NERFbin","lcfbin","sfpbin", | ||
"EogNERF","Eoglcf","Eogsfp"))==FALSE], min), 2, sum)) - sum(apply(tapply(temp2$Freq, temp2[,c("Scenario","Age")], mean), 2, min)) | "EogNERF","Eoglcf","Eogsfp"))==FALSE], min), 2, sum)) - sum(apply(tapply(temp2$Freq, temp2[,c("Scenario","Age")], mean), 2, min)) | ||
mean(apply(tapply(temp2$Eoglcf, temp2[,(colnames(temp2)%in%c("Freq","Scenario","tNERF","tlcf","tsfp","NERFbin","lcfbin","sfpbin", | EVXPI.lcf <- mean(apply(tapply(temp2$Eoglcf, temp2[,(colnames(temp2)%in%c("Freq","Scenario","tNERF","tlcf","tsfp","NERFbin","lcfbin","sfpbin", | ||
"EogNERF","Eoglcf","Eogsfp"))==FALSE], min), 2, sum)) - sum(apply(tapply(temp2$Freq, temp2[,c("Scenario","Age")], mean), 2, min)) | "EogNERF","Eoglcf","Eogsfp"))==FALSE], min), 2, sum)) - sum(apply(tapply(temp2$Freq, temp2[,c("Scenario","Age")], mean), 2, min)) | ||
mean(apply(tapply(temp2$Eogsfp, temp2[,(colnames(temp2)%in%c("Freq","Scenario","tNERF","tlcf","tsfp","NERFbin","lcfbin","sfpbin", | EVXPI.sfp <- mean(apply(tapply(temp2$Eogsfp, temp2[,(colnames(temp2)%in%c("Freq","Scenario","tNERF","tlcf","tsfp","NERFbin","lcfbin","sfpbin", | ||
"EogNERF","Eoglcf","Eogsfp"))==FALSE], min), 2, sum)) - sum(apply(tapply(temp2$Freq, temp2[,c("Scenario","Age")], mean), 2, min)) | "EogNERF","Eoglcf","Eogsfp"))==FALSE], min), 2, sum)) - sum(apply(tapply(temp2$Freq, temp2[,c("Scenario","Age")], mean), 2, min)) | ||
VOI <- data.frame(VOI = c("Total VOI (EVPI)", "Narcolepsy ERF", "Fraction of lab-confirmed cases", "P(swine flu|non-immune)"), value = c(EVPI, EVXPI.NERF, EVXPI.lcf, EVXPI.sfp)) | |||
colnames(VOI)[2] <- "Value in DALYs" | |||
print(xtable(VOI), type = 'html') | |||
#vacscenario <- outcome() | #vacscenario <- outcome() | ||
| Line 389: | Line 427: | ||
#temp <- as.data.frame(as.table(tapply(fout$Result, fout[,c("obs","Scenario")], sum))) | #temp <- as.data.frame(as.table(tapply(fout$Result, fout[,c("obs","Scenario")], sum))) | ||
#ggplot(temp, aes(x = Freq, fill = Scenario)) + geom_density(alpha = 0.2) | #ggplot(temp, aes(x = Freq, fill = Scenario)) + geom_density(alpha = 0.2) | ||
</rcode> | |||
==See also== | |||
* [http://www.academia.edu/527623/Online_authority_communication_during_an_epidemic_A_Finnish_example Päivi Tirkkonen, Vilma Luoma-aho]: Online authority communication during an epidemic: A Finnish example | |||
* [http://www.journalsleep.org/ViewAbstract.aspx?pid=27949 About narcolepsy in Journal Sleep] | |||
* [http://www.thl.fi/thl-client/pdfs/dce182fb-651e-48a1-b018-3f774d6d1875 Interim Report of the National Narcolepsy Task Force] in Finland | |||
* [http://www.who.int/vaccine_safety/topics/influenza/pandemic/h1n1_safety_assessing/narcolepsy_statement/en/index.html WHO Statement on narcolepsy and vaccination] | |||
* [http://www.thl.fi/en_US/web/en/pressrelease?id=26352 THL press release about narcolepsy, 1 Sept 2011] | |||
Scientific publications in Pubmed: | |||
[http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3248605] | |||
[http://www.ncbi.nlm.nih.gov/pubmed/22172962] | |||
[http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3192001] | |||
[http://www.ncbi.nlm.nih.gov/pubmed/21534891] | |||
[http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3099488] | |||
[http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2954689] | |||
[http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2954687] | |||
[http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2954688] | |||
==Keywords== | |||
Swine flu, influenza, vaccination, narcolepsy, risk perception, risk comparison | |||
==References== | |||
<references/> | |||
==Related files== | |||
{{mfiles}} | |||
{{eracedu}} | |||
Latest revision as of 12:18, 26 August 2014
| Moderator:Teemu R (see all) |
|
|
| Upload data
|
This assessment was completed in 2011, it is no longer actively updated.
| Main message: |
|---|
| Question:
What was the overall health impact of the H1N1 (swine flu) vaccination in Finland in 2009-2010? Given current knowledge, which was the better decision between vaccinating as happened versus vaccinating no-one versus not vaccinating the population aged 5-19? Given current (2011) knowledge, the decision to vaccinate the whole population was the best decision even when narcolepsy is included in the assessment. Results of the Value of information analysis suggest that further knowledge about the uncertain variables considered very likely would not have changed the decision. |
This assessment is about the total health effects the 2009 swine flu pandemic. It utilizes data from the Infectious disease registry (TTR) maintained by THL, the National narcolepsy task force report from 31.1.2011 and WHO. The current model is a simplification with no time dimension. The assessment has been evaluated. Evaluation results are displayed on the discussion page
Scope
- What was the overall health impact of the H1N1 vaccination in Finland in 2009-2010?
- Given current knowledge, which was the better decision between vaccinating as happened versus vaccinating no-one versus not vaccinating the population aged 5-19?
- Monetary impact is not considered.
Participants
- THL: Virpi Kollanus, M.Sc., Marko Tainio, Ph.D., Mikko V. Pohjola, M.Tech.Sci., Jouni T. Tuomisto, M.D., Dr.Med.Sci., Teemu Rintala
- This is an open assessment. Anyone can participate.
Result
{{#opasnet_base_link:Op_en4926}}
Results

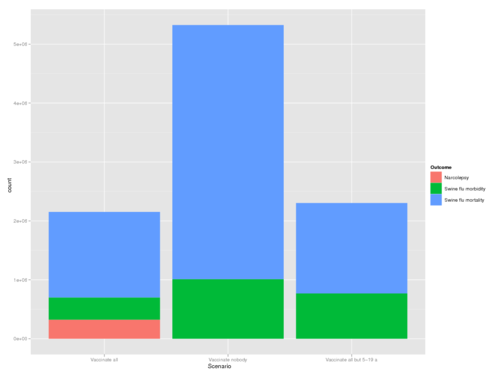
- From initial results it would appear like swine flu is more significant than narcolepsy in terms of DALYs.
- Vaccinating as planned would result in approximately 1850 DALYs due to swine flu and narcolepsy combined.
- Vaccinating no-one would result in approximately 4400 DALYs due to swine flu.
- Vaccinating everyone but people aged 5-19 would result in about a total of 2000 DALYs.
- Probability of swine flu variable is revealed by both the sensitivity- and Value of information-analyses to have the most impact on the outcome.
- The VOI analysis also reveals that further knowledge about the uncertain variables in the model is only worth up to ~80 DALYs, when considering the decision by age group, and less than 1 DALY when considering the decision as on/off as defined in the decision variable above. Which is only a small fraction of the total DALYs.
- Suggested statement: Pandemrix should not be used any more anywhere because its narcolepsy risk is too high.
- Resolution: Not accepted. Pandemrix is still an effective and safe vaccine. However, due to precautionary reasons, other alternatives should be used when available, because the occurrence of narcolepsy is not understood. R↻
Conclusions
Given current knowledge, the decision to vaccinate the whole population was the best decision. Results of the Value of information analysis suggest that further knowledge about the uncertain variables considered very likely would not have changed the decision. The total impact of the swine flu pandemic and related narcolepsy cases in Finland in terms of DALYs is slightly smaller than that of radon (~6700 DALYs yearly) and slightly larger than that of moisture damage (~650 DALYs yearly) for instance. It should be noted that only three variables had their uncertainty taken into account, although they should represent the major uncertainties present. Also, herd immunity is assumed not to affect the probability of a non immune subject to catch swine flu, this results in an underestimation of the number of swine flu cases in scenarios where the vaccination coverage is less than what was observed.
Rationale

- Decisions
- Vaccination decision
- Vaccinate everyone (observed vaccination coverage)
- Vaccinate no-one (0 vaccination coverage)
- Variables
- H1N1 vaccination coverage in Finland
- ERF of H1N1 vaccination on Narcolepsy
- Assumed lognormally distributed
- A(H1N1)v immunity in the Finnish population
- P(immune) = 1 - P(not vaccinated) * P(no base immunity)
- Population of Finland
- Disability weights
- DALY weight of narcolepsy equals roughly that of epilepsy (0.065)
- DALY weight of swine flu for the duration of the disease ~ 0.5 (~ 0.007 DALYs per case when adjusted for duration)
- Life expectancy by age groups in Finland[1]
- Probability of catching swine flu given subject is not immune
- Estimated from data available (population, total immunity, number of cases) by fitting the number of cases to a poisson distribution and calculating probability from the mean estimate by dividing by the non-immune population
- Fraction of all cases represented by lab confirmed cases (which we have data on)
- Estimated as beta-distributed with mean of 0.2 and some sd
- Probability of death due to swine flu given a subject has swine flu and belongs to a risk group
- Estimated from data available
- Assumed all deaths will be lab confirmed cases
- Assumed that all deaths belonged to a risk group (had some base condition)
- Fraction of population belonging to a risk group
- Arbitrary values; trying to account for kids of age <1 and old folks with heart conditions etc.
- Duration of swine flu
- Assumed to be flat 5 days (mildly incapacitated for this duration)
- Narcolepsy in Finland
- AH1N1 cases in Finland
- Indicators
- DALYs from narcolepsy caused by vaccination
- DALYs from having swine flu
- DALYs from deaths caused by swine flu
R code
- Basic model
- Uncertainties of ERF of vaccine on narcolepsy, fraction of all cases represented by lab confirmed cases and probability of catching swine flu are implemented.
See also
- Päivi Tirkkonen, Vilma Luoma-aho: Online authority communication during an epidemic: A Finnish example
- About narcolepsy in Journal Sleep
- Interim Report of the National Narcolepsy Task Force in Finland
- WHO Statement on narcolepsy and vaccination
- THL press release about narcolepsy, 1 Sept 2011
Scientific publications in Pubmed: [2] [3] [4] [5] [6] [7] [8] [9]
Keywords
Swine flu, influenza, vaccination, narcolepsy, risk perception, risk comparison
References
Related files
<mfanonymousfilelist></mfanonymousfilelist>

|

|