Goherr: Fish consumption study: Difference between revisions
| Line 689: | Line 689: | ||
cat("Average fish consumption in subgroups\n") | cat("Average fish consumption in subgroups\n") | ||
oprint(tmp) | oprint(tmp) | ||
</rcode> | |||
==== Regression analysis, ANOVA, and Tukey ==== | |||
{{argument|relat1=relevant attack|selftruth1=true|id=arg7420|type=|content=Work in progress.|sign=--[[User:Jouni|Jouni]] ([[User talk:Jouni|talk]]) 10:39, 8 August 2018 (UTC)}} | |||
<rcode graphics=1> | |||
# This is code Op_en7749/surveyjsp on page [[Goherr: Fish consumption study]] | |||
# The code produces amount esimates (jsp ovariable) directly from data rather than bayesian model. | |||
library(OpasnetUtils) | |||
jsp <- Ovariable( | |||
"jsp", | |||
dependencies = data.frame(Name="survey1", Ident="Op_en7749/preprocess2"), # [[Goherr: Food cunsumption study]] | |||
formula = function(...) { | |||
require(reshape2) | |||
sur <- survey1[c(157,1,3,158,162:178,81,82,131,132)] # Removed, not needed:16,29,30? | |||
#colnames(sur) | |||
#[1] "Row" "Country" "Gender" | |||
#[4] "Ages" "Baltic.salmon.n" "How.often.BS.n" | |||
#[7] "How.much.BS.n" "How.often.side.BS.n" "How.much.side.BS.n" | |||
#[10] "Better.availability.BS.n" "Less.chemicals.BS.n" "Eat.BH.n" | |||
#[13] "How.often.BH.n" "How.much.BH.n" "How.often.side.BH.n" | |||
#[16] "How.much.side.BH.n" "Better.availability.BH.n" "Less.chemicals.BH.n" | |||
#[19] "Eatfish" "Eatsalm" "Eatherr" | |||
#[22] "Recommended.BS" "Not.recommended.BS" "Recommended.BH" | |||
#[25] "Not.recommended.BH" | |||
# Make sure that Row is kept separate from Iter because in the sampling version they are different. | |||
# sur contained columns Eat.fish, How.often.fish, Eat.salmon. Are these needed, as all other questions are melted? No | |||
# Columns 5-7 removed, so colnames list above does not match. | |||
colnames(sur)[5] <- "Eat.BS" | |||
colnames(sur) <- gsub("\\.n","",colnames(sur)) | |||
sur[22:25] <- sapply(sur[22:25], as.numeric) | |||
sur <- melt( | |||
sur, | |||
id.vars=c(1:4), | |||
variable.name="Question", | |||
value.name="Result" | |||
) | |||
sur$Fish <- ifelse(grepl("BH",sur$Question),"Herring","Salmon") | |||
sur$Question <- gsub("\\.BS","",sur$Question) | |||
sur$Question <- gsub("\\.BH","",sur$Question) | |||
########## If the missing values are not adjusted, they drop out in the next stage. | |||
if(TRUE) { | |||
# The adjustments below probably should go to the preprocess2 code. | |||
sur$Result[sur$Question %in% c( | |||
"Better.availability", | |||
"Less.chemicals", | |||
"Recommended", | |||
"Not.recommended" | |||
) & is.na(sur$Result)] <- 4 # If missing --> no change | |||
sur$Result[is.na(sur$Result)] <- 1 # Replace missing values with 1. That will produce 0 g/d. | |||
} | |||
return(Ovariable( | |||
output = sur, | |||
marginal = colnames(sur) %in% c("Fish", "Iter", "Question") | |||
)) | |||
} | |||
) | |||
objects.store(jsp) | |||
cat("Ovariable jsp with actual survey data: every respondent is kept in data without sampling.\n") | |||
############################ | |||
library(OpasnetUtils) | |||
library(ggplot2) | |||
openv.setN(10) | |||
objects.latest("Op_en7749",code_name="initiate") | |||
effinfo <- 0 | |||
effrecomm <- 0 | |||
amount <- EvalOutput(amount) | |||
result(amount)[result(amount)==0] <- 0.1 | |||
dat <- merge(survey1,amount@output) | |||
#table 2: statistics of the survey population in each country (n, female %, education, purchasing power) | |||
round(t(data.frame( | |||
n = tapply(rep(1,nrow(survey1)), survey1$Country,sum), | |||
Females = tapply(survey1$Gender=="Female", survey1$Country,mean)*100, | |||
Old = tapply(survey1$Ages==">45", survey1$Country,mean)*100, | |||
PurcVlow = tapply(survey1$Purchasing.power=="Very low", survey1$Country,mean)*100, | |||
PurcLow = tapply(survey1$Purchasing.power=="Low", survey1$Country,mean)*100, | |||
PurcSuf = tapply(survey1$Purchasing.power=="Sufficient", survey1$Country,mean)*100, | |||
PurcGood = tapply(survey1$Purchasing.power=="Good", survey1$Country,mean)*100, | |||
PurcVgood = tapply(survey1$Purchasing.power=="Very good", survey1$Country,mean)*100, | |||
PurcExc = tapply(survey1$Purchasing.power=="Excellent", survey1$Country,mean)*100, | |||
EducPri = tapply(survey1$Education=="Primary education", survey1$Country,mean)*100, | |||
EducSec = tapply(survey1$Education=="Secondary education (gymnasium, vocational school or similar)", survey1$Country,mean)*100, | |||
EducCol = tapply(survey1$Education=="Lower level college education or similar", survey1$Country,mean)*100, | |||
EducHig = tapply(survey1$Education=="Higher level college education or similar", survey1$Country,mean)*100 | |||
))) | |||
# These should be corrected to preprocess2. NOT ordered. | |||
dat$Country <- factor(dat$Country, ordered=FALSE) | |||
dat$Ages <- factor(dat$Ages, ordered=FALSE) | |||
dat$Gender <- factor(dat$Gender, ordered=FALSE) | |||
ggplot(dat, aes(x=amountResult, weight=1, color=Country))+stat_ecdf()+facet_wrap(~Fish)+ | |||
scale_x_log10() | |||
out <- data.frame() | |||
for(i in unique(dat$Fish)) { | |||
# for(j in unique(dat$Country)){ | |||
for(k in unique(dat$Iter)) { | |||
res <- lm(log10(amountResult) ~ Gender + Ages + Country, data=dat[ | |||
dat$Iter==k & dat$Fish==i,]) # & dat$Country==j | |||
out <- rbind(out, data.frame( | |||
summary(res)[[4]], | |||
Fish=i, | |||
Country=j, | |||
Iter=k, | |||
Topic=rownames(summary(res)[[4]]) | |||
)) | |||
} | |||
# } | |||
} | |||
ggplot(out, aes(x=paste(Fish, Country), y = Pr...t.., colour = Topic))+geom_jitter() | |||
###################### The following is directly from AAH study. Adjust for this study. | |||
################################# ANOVA AND TUKEY'S TEST | |||
#### test if means differ among gender, age or expertise within aah_average and nonaah_average and make a table | |||
# make a list to store statistical results | |||
varnames <- c("aah_average", "nonaah_average", "nonaah.average12", "Social.average", "Enviro.average") | |||
explnames <- c("Gender", "Age", "Expertise", "Expertise2", "Pub.all.peer", "Pub.hum.peer", "Pub.all.popular", | |||
"Pub.hum.popular", "Familiarity", "Teaching") | |||
M <- matrix(data=NA, nrow=length(explnames), ncol=length(varnames)) | |||
colnames(M) <- varnames | |||
rownames(M) <- explnames | |||
res.all <- apply(M, 1, as.list) | |||
# run the analyses of variance | |||
for(i in explnames) { | |||
for(j in varnames) { | |||
subset <- survey1[,c(i,j)] | |||
colnames(subset) <- c("Explanation", "Variable") | |||
# subset$Variable <- as.numeric(subset$Variable) | |||
res.aov <- aov(Variable ~ Explanation, data=subset) | |||
res.tukey <- TukeyHSD(res.aov) | |||
res.all[[i]][[j]][1:2] <- list(res.aov, res.tukey) | |||
} | |||
} | |||
oprint(res.all) | |||
####################### Means and SDs of average indices by several determinants | |||
cat("Means and SDs of average indices by several determinants.\n") | |||
meantable <- function(col) { | |||
out <- aggregate( | |||
survey1[varnames], | |||
by = survey1[col], | |||
FUN = function(x) { | |||
paste( | |||
sprintf("%.2f", mean(as.numeric(x), na.rm = TRUE)), " (", | |||
sprintf("%.2f", sd(as.numeric(x), na.rm = TRUE)), ")", sep="" | |||
) | |||
} | |||
) | |||
colnames(out)[1] <- "Subgroup" | |||
out <- rbind( | |||
data.frame( # significant p values summarised | |||
Subgroup = col, | |||
sapply( | |||
varnames, | |||
FUN = function(x) { | |||
pees <- res.all[[col]][[x]][2][[1]][[1]][,4] # p values from ANOVA/Tukey | |||
pees <- pees[pees < 0.05] | |||
pees <- paste( | |||
paste( | |||
names(pees), | |||
cut(pees, breaks = c(-1,0.001,0.01, 0.05), labels = c("***", "**", "*")) | |||
), collapse = " " | |||
) | |||
}, | |||
simplify = FALSE | |||
) | |||
), | |||
out # means and sd's | |||
) | |||
return(out) | |||
} | |||
means <- rbind( | |||
meantable("Expertise"), | |||
meantable("Familiarity"), | |||
meantable("Gender"), | |||
meantable("Age"), | |||
meantable("Pub.hum.peer"), | |||
meantable("Teaching") | |||
) | |||
oprint(means) | |||
############################################# ANOVA TUKEY WITH INTERACTIONS | |||
res.aov <- aov(aah_average ~ Teaching*Expertise, data=survey1) | |||
res.tukey <- TukeyHSD(res.aov) | |||
oprint(list(res.aov, res.tukey)) | |||
res.aov <- aov(aah_average ~ Familiarity*Expertise, data=survey1) | |||
res.tukey <- TukeyHSD(res.aov) | |||
oprint(list(res.aov, res.tukey)) | |||
############################################# TUKEY INTERACTIONS IN NICE TABLES | |||
res.aov <- aov(aah_average ~ Familiarity*Expertise, data=survey1) | |||
res.tukey <- TukeyHSD(res.aov) | |||
out <- res.tukey[[3]][order(dimnames(res.tukey[[3]])[[1]]),] | |||
test <- lapply( # Split the comparison items | |||
strsplit(dimnames(out)[[1]], split="-"), | |||
FUN = function(x) strsplit(x, split = ":") | |||
) | |||
oprint(out[sapply(test, FUN = function(x) x[[1]][1] == x[[2]][1]),]) # Matching Familiarity | |||
oprint(out[sapply(test, FUN = function(x) x[[1]][2] == x[[2]][2]),]) # Matching Expertise | |||
</rcode> | </rcode> | ||
Revision as of 10:39, 8 August 2018
| [show] This page is a study.
The page identifier is Op_en7749 |
|---|
Question
How Baltic herring and salmon are used as human food in Baltic sea countries? Which determinants affect on people’s eating habits of these fish species?
Answer
Original questionnaire analysis results
- 13.3.2017 ----#: . These should be presented somewhere --Arja (talk) 07:39, 26 April 2017 (UTC) (type: truth; paradigms: science: comment)
Consumption amount estimates
- Model run 21.4.2017 [1] first distribution
- Model run 18.5.2017 with modelled data; with direct survey data
Rationale
Survey of eating habits of Baltic herring and salmon in Denmark, Estonia, Finland and Sweden has been done in September 2016 by Taloustutkimus oy. Content of the questionnaire can be accessed in Google drive. The actual data can be found from the link below (see Data).
Data
Questionnaire
Original datafile File:Goherr fish consumption.csv.
| [show]Show details |
|---|
Assumptions
The following assumptions are used:
| Obs | Variable | Value | Unit | Result | Description |
|---|---|---|---|---|---|
| 1 | freq | 1 | times /a | 0 | Never |
| 2 | freq | 2 | times /a | 0.5 - 0.9 | less than once a year |
| 3 | freq | 3 | times /a | 2 - 5 | A few times a year |
| 4 | freq | 4 | times /a | 12 - 36 | 1 - 3 times per month |
| 5 | freq | 5 | times /a | 52 | once a week |
| 6 | freq | 6 | times /a | 104 - 208 | 2 - 4 times per week |
| 7 | freq | 7 | times /a | 260 - 364 | 5 or more times per week |
| 8 | amdish | 1 | g /serving | 20 - 70 | 1/6 plate or below (50 grams) |
| 9 | amdish | 2 | g /serving | 70 - 130 | 1/3 plate (100 grams) |
| 10 | amdish | 3 | g /serving | 120 - 180 | 1/2 plate (150 grams) |
| 11 | amdish | 4 | g /serving | 170 - 230 | 2/3 plate (200 grams) |
| 12 | amdish | 5 | g /serving | 220 - 280 | 5/6 plate (250 grams) |
| 13 | amdish | 6 | g /serving | 270 - 400 | full plate (300 grams) |
| 14 | amdish | 7 | g /serving | 400 - 550 | overly full plate (500 grams) |
| 15 | ingredient | fraction | 0.1 - 0.3 | Fraction of fish in the dish | |
| 16 | amside | 1 | g /serving | 20 - 70 | 1/6 plate or below (50 grams) |
| 17 | amside | 2 | g /serving | 70 - 130 | 1/4 plate (100 grams) |
| 18 | amside | 3 | g /serving | 120 - 180 | 1/2 plate (150 grams) |
| 19 | amside | 4 | g /serving | 170 - 230 | 2/3 plate (200 grams) |
| 20 | amside | 5 | g /serving | 220 - 280 | 5/6 plate (250 grams) |
| 21 | change | 1 | fraction | -1 - -0.8 | Decrease it to zero |
| 22 | change | 2 | fraction | -0.9 - -0.5 | Decrease it to less than half |
| 23 | change | 3 | fraction | -0.6 - -0.1 | Decrease it a bit |
| 24 | change | 4 | fraction | 0 | No effect |
| 25 | change | 5 | fraction | 0.1 - 0.6 | Increase it a bit |
| 26 | change | 6 | fraction | 0.5 - 0.9 | Increase it over by half |
| 27 | change | 7 | fraction | 0.8 - 1.3 | Increase it over to double |
| 28 | change | 8 | fraction | -0.3 - 0.3 | Don't know |
Preprocessing
This code is used to preprocess the original questionnaire data from the above .csv file and to store the data as a usable variable to Opasnet base. The code stores a data.frame named survey.
- Model run 11.7.2018 [2]
Analyses
- Sketches about modelling determinants of eating (spring 2018) [3]
Figures for the first manuscript
Regression analysis, ANOVA, and Tukey
arg7420: . Work in progress. --Jouni (talk) 10:39, 8 August 2018 (UTC) (type: ; paradigms: science: relevant attack)
Descriptive statistics
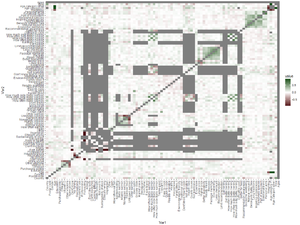
Model must contain predictors such as country, gender, age etc. Maybe we should first study what determinants are important? Model must also contain determinants that would increase or decrease fish consumption. This should be conditional on the current consumption. How? Maybe we should look at principal coordinates analysis with all questions to see how they behave.
Also look at correlation table to see clusters.
Some obvious results:
- If reports no fish eating, many subsequent answers are NA.
- No vitamins correlates negatively with vitamin intake.
- Unknown salmon correlates negatively with the types of salmon eaten.
- Different age categories correlate with each other.
However, there are also meaningful negative correlations:
- Country vs allergy
- Country vs Norwegian salmon and Rainbow trout
- Country vs not traditional.
- Country vs recommendation awareness
- Allergy vs economic wellbeing
- Baltic salmon use (4 questions) vs Don't like taste and Not used to
- All questions between Easy to cook ... Traditional dish
Meaningful positive correlations:
- All questions between Baltic salmon ... Rainbow trout
- How often Baltic salmon/herring/side salmon/side herring
- How much Baltic salmon/herring/side salmon/side herring
- Better availability ... Recommendation
- All questions between Economic wellbeing...Personal aims
- Omega3, Vitamin D, and Other vitamins
Model runs
- Model run 13.3.2017
- Model run 21.4.2017 [8] old code from Answer merged to this code and debugged
Bayes model
- Model run 3.3.2017. All variables assumed independent. [9]
- Model run 3.3.2017. p has more dimensions. [10]
- Model run 25.3.2017. Several model versions: strange binomial+multivarnormal, binomial, fractalised multivarnormal [11]
- Model run 27.3.2017 [12]
- Other models except multivariate normal were archived and removed from active code 29.3.2017.
- Model run 29.3.2017 with raw data graphs [13]
- Model run 29.3.2017 with salmon and herring ovariables stored [14]
- Model run 13.4.2017 with first version of coordinate matrix and principal coordinate analysis [15]
- Model run 20.4.2017 [16] code works but needs a safety check against outliers
- Model run 21.4.2017 [17] some model results plotted
- Model run 21.4.2017 [18] ovariables produced by the model stored.
- Model run 18.5.2017 [19] small updates
- 13.2.2018 old model run but with new Opasnet [20]
Initiate ovariables
Amount estimated from a bayesian model.
- Model run 24.5.2017 [21]
Amount estimates directly from data rather than from a bayesian model.
- Initiation run 18.5.2017 [22]
- Initiation run 24.2.2018: sampling from survey rather than each respondent once [23]
Initiate other ovariables
- Code stores ovariables assump, often, much, oftenside, muchside, amount.
- Model run 19.5.2017 [24]
- Initiation run 24.5.2017 without jsp [25]
- Model run 8.6.2017 [26]
Dependencies
The survey data will be used as input in the benefit-risk assessment of Baltic herring and salmon intake, which is part of the WP5 work in Goherr-project.
See also
- Useful information about Wishart distribution and related topics:
Keywords
References
Related files