Epidemiological modelling: Difference between revisions
(progression class added) |
(Laskenta: nyt sama koodi kuin suomenk puolella) |
||
| Line 33: | Line 33: | ||
== Computation == | |||
The following program illustrates the working of the replacement model. In its current implementation the code allows the user to specify 4 vaccine compositions and then displays the predicted IPD cases in Finland per year corresponding to these vaccines. The results are shown by serotype and by age category (<5 and 5+ year olds). Possible choices for vaccine compositions are: PCV10, PCV13, no vaccination and user specified serotype composition. The program is based on the code in File S1 of Nurhonen and Auranen, 2014. | |||
Instructions for user: Choose the desired vaccine compositions from the list below and then press "Run code". The results will be displayed on a separate tab. The default choices are PCV10 and PCV13. | |||
<rcode embed= | <rcode embed=0 graphics=1 variables=" | ||
name:vac|description:Please choose the vaccines to be compared:|type:checkbox|options: | |||
' | 'PCV10';PCV-10; | ||
' | 'PCV13';PCV-13; | ||
'16';16;'20';20;'22';22;'23A'; | 'No_vaccination';No vaccination| | ||
default:'19F';'23F';'6B';'14';'9V';'4';'18C';'1';'7' | default:'PCV10';'PCV13'| | ||
"> | category:Skenaariot| | ||
name:custom_vac|description:Do you want to specify another vaccine composition?|type:selection|options: | |||
FALSE;No; | |||
TRUE;Yes| | |||
default:FALSE| | |||
name:vac_user|description:Choose the serotypes for the user defined vaccine composition|type:checkbox|options: | |||
'1';1; | |||
'3';3; | |||
'4';4; | |||
'6A';6A; | |||
'6B';6B; | |||
'6C';6C; | |||
'7';7F; | |||
'8';8; | |||
'9N';9N; | |||
'9V';9V; | |||
'10';10; | |||
'11';11; | |||
'12';12; | |||
'14';14; | |||
'15';15; | |||
'16';16; | |||
'18C';18C; | |||
'19A';19A; | |||
'19F';19F; | |||
'20';20; | |||
'22';22; | |||
'23A';23A; | |||
'23F';23F; | |||
'33';33; | |||
'35';35; | |||
'38';38; | |||
'Oth';Other| | |||
default:'19F';'23F';'6B';'14';'9V';'4';'18C';'1';'7'| | |||
category:Oma rokote| | |||
category_conditions:custom_vac;TRUE" | |||
> | |||
library(OpasnetUtils) | library(OpasnetUtils) | ||
library(ggplot2) | library(ggplot2) | ||
#vacc1 <- vac | |||
#vacc2 <- custom_vac | |||
if(custom_vac) { | |||
vac <- c(vac, "NewVaccine") | |||
} | |||
if (length(vac) == 0) stop("Mitään skenaariota ei valittu") | |||
user_args <- list( | |||
Scenario = vac | |||
) | |||
# Ulkoinen säilö datalle jollain sivulla? | |||
temp <- data.frame( | |||
Vaccine = rep(c("PCV10", "PCV13"), c(9, 12)), | |||
Serotype = c('19F', '23F', '6B', '14', '9V', '4', '18C', '1', '7', | |||
'19F', '23F', '6B', '14', '9V', '4', '18C', '1', '7', '3', '6A', '19A' | |||
) | |||
) | |||
user_args$Vaccines <- temp[temp$Vaccine %in% user_args$Scenario, ] | |||
if(custom_vac) { | |||
user_args$Vaccines <- rbind( | |||
user_args$Vaccines, | |||
data.frame(Vaccine = "NewVaccine", Serotype = vac_user) | |||
) | |||
} | |||
#if(!exists("servac_user")) servac_user <- c('19F', '23F', '6B', '14', '9V', '4', '18C', '1', '7') | |||
objects.latest("Op_fi4305", code_name = "alusta") # [[Pneumokokkirokote]] | objects.latest("Op_fi4305", code_name = "alusta") # [[Pneumokokkirokote]] | ||
| Line 58: | Line 129: | ||
## The 0 entries in IPD and carriage data are replaced by small values. | ## The 0 entries in IPD and carriage data are replaced by small values. | ||
serotypes<-c( | serotypes<-c( | ||
"19F", "23F", "6B", "14", "9V", "4", "18C", "1", "7", | "19F", "23F", "6B", "14", "9V", "4", "18C", "1", "7", | ||
"6A", "19A", "3", "8", "9N", "10", "11", "12", "15", | "6A", "19A", "3", "8", "9N", "10", "11", "12", "15", | ||
"16", "20", "22", "23A", "33", "35", "38", "6C", "Oth") | "16", "20", "22", "23A", "33", "35", "38", "6C", "Oth") | ||
car_under5<-c( | car_under5<-c( | ||
156030, 156030, 126990, 41200, 22290, 12830, 10130, 10, 14180, | 156030, 156030, 126990, 41200, 22290, 12830, 10130, 10, 14180, | ||
54940, 24320, 12160, 1350, 20940, 4050, 72270, 10, 33100, | 54940, 24320, 12160, 1350, 20940, 4050, 72270, 10, 33100, | ||
3380, 1350, 12160, 3380, 680, 30400, 4050, 27470, 24320 ) | 3380, 1350, 12160, 3380, 680, 30400, 4050, 27470, 24320 ) | ||
car_over5<-c( | car_over5<-c( | ||
168100, 314800, 256700, 209800, 114100, 62500, 200700, 100, 100, | 168100, 314800, 256700, 209800, 114100, 62500, 200700, 100, 100, | ||
158800, 54900, 30800, 8800, 8800, 20800, 97700, 100, 100, | 158800, 54900, 30800, 8800, 8800, 20800, 97700, 100, 100, | ||
191900, 25200, 72500, 22000, 100, 71300, 100, 79400, 330100 ) | 191900, 25200, 72500, 22000, 100, 71300, 100, 79400, 330100 ) | ||
ipd_under5<-c( | ipd_under5<-c( | ||
7.78, 7.88, 24.39, 20.76, 2.91, 2.91, 6.64, 0.31, 3.02, | 7.78, 7.88, 24.39, 20.76, 2.91, 2.91, 6.64, 0.31, 3.02, | ||
3.94, 9.88, 1.25, 0.10, 0.83, 0.41, 0.42, 0.21, 1.98, | 3.94, 9.88, 1.25, 0.10, 0.83, 0.41, 0.42, 0.21, 1.98, | ||
0.21, 0.01, 0.93, 0.10, 0.42, 0.31, 0.42, 0.01, 0.73 ) | 0.21, 0.01, 0.93, 0.10, 0.42, 0.31, 0.42, 0.01, 0.73 ) | ||
ipd_over5<-c( | ipd_over5<-c( | ||
28.51, 53.72, 29.53, 99.43, 43.07, 76.99, 24.39, 6.58, 46.88, | 28.51, 53.72, 29.53, 99.43, 43.07, 76.99, 24.39, 6.58, 46.88, | ||
17.42, 20.54, 55.04, 11.21, 25.20, 6.28, 12.76, 13.89, 9.18, | 17.42, 20.54, 55.04, 11.21, 25.20, 6.28, 12.76, 13.89, 9.18, | ||
4.73, 3.29, 29.03, 4.40, 5.64, 12.41, 1.43, 5.50, 11.20 ) | 4.73, 3.29, 29.03, 4.40, 5.64, 12.41, 1.43, 5.50, 11.20 ) | ||
## Combine the data into 2 matrices of dimension 27*2: | ## Combine the data into 2 matrices of dimension 27*2: | ||
| Line 112: | Line 183: | ||
## replacement is 50% and the current composition includes the PCV7 serotypes. | ## replacement is 50% and the current composition includes the PCV7 serotypes. | ||
opt<-OptimalSequence(IPD,Car, VT_rows=pcv7rows,Excluded_rows=length(serotypes), | opt<-OptimalSequence(IPD,Car, VT_rows=pcv7rows,Excluded_rows=length(serotypes), | ||
p=0.5,HowmanyAdded=17) | p=0.5,HowmanyAdded=17) | ||
| Line 126: | Line 197: | ||
Car@data <- Car@data[Car@data$Observation == "Carrier" , colnames(Car@data) != "Observation"] | Car@data <- Car@data[Car@data$Observation == "Carrier" , colnames(Car@data) != "Observation"] | ||
servac <- Ovariable("servac", data = data.frame( | servac <- merge(data.frame(Vaccine = user_args$Scenario), data.frame(Serotype = serotypes)) | ||
servac <- merge( | |||
data.frame(user_args$Vaccines, Result = 1), | |||
servac, | |||
all.y = TRUE | |||
) | |||
)) | servac$Result <- as.numeric(!is.na(servac$Result)) | ||
)) | servac <- Ovariable( | ||
"servac", | |||
data = servac | |||
) | |||
#data.frame( | |||
#Vaccine = rep(c("Current", "New"), each = length(serotypes)), | |||
#Serotype = serotypes, | |||
#Result = as.numeric(c( | |||
# serotypes %in% c("19F", "23F", "6B", "14", "9V", "4", "18C", "1", "7"), | |||
# serotypes %in% servac_user | |||
# )) | |||
#)) | |||
p_user<-q_user<-adultcarriers<-1 | p_user<-q_user<-adultcarriers<-1 | ||
| Line 148: | Line 230: | ||
if (1==0) { | if (1==0) { | ||
cat("servac\n") | cat("servac\n") | ||
oprint(summary(servac)) | oprint(summary(servac)) | ||
cat("Number of carriers\n") | cat("Number of carriers\n") | ||
oprint(summary(VacCar)) | oprint(summary(VacCar)) | ||
cat("Incidence of invasive pneumococcal disease.\n") | cat("Incidence of invasive pneumococcal disease.\n") | ||
oprint(summary(VacIPD)) } | oprint(summary(VacIPD)) } | ||
| Line 163: | Line 245: | ||
if (1==0) {ggplot(VacCar@output, aes(x = Serotype, weight = result(VacCar) / N, fill = Vaccine)) + geom_bar(position = "dodge") + theme_gray(base_size = 24) + | if (1==0) {ggplot(VacCar@output, aes(x = Serotype, weight = result(VacCar) / N, fill = Vaccine)) + geom_bar(position = "dodge") + theme_gray(base_size = 24) + | ||
labs(title = "Carriers", y = "Number of carriers in Finland") } | |||
ggplot(VacIPD@output, aes(x = Serotype, weight = result(VacIPD) / N, fill = Vaccine)) + geom_bar(position = "dodge") + theme_gray(base_size = 24) + | ggplot(VacIPD@output, aes(x = Serotype, weight = result(VacIPD) / N, fill = Vaccine)) + geom_bar(position = "dodge") + theme_gray(base_size = 24) + | ||
labs(title = "Incidence of invasive pneumococcal disease", y = "Number of cases per year") | |||
ggplot(VacIPD@output, aes(x = Vaccine, weight = result(VacIPD) / N, fill = Age)) + geom_bar(position = "stack") + theme_gray(base_size = 24) + | ggplot(VacIPD@output, aes(x = Vaccine, weight = result(VacIPD) / N, fill = Age)) + geom_bar(position = "stack") + theme_gray(base_size = 24) + | ||
labs(title = "Incidence of invasive pneumococcal disease", y = "Number of cases per year") | |||
</rcode> | </rcode> | ||
=== | === Funktioiden alustus === | ||
<rcode name="alusta" label=" | <rcode name="alusta" label="Alusta funktiot" embed=1> | ||
library(OpasnetUtils) | library(OpasnetUtils) | ||
| Line 403: | Line 485: | ||
objects.store(Vaccination, NextVT, OptimalSequence, OptimalVacc, VacCar, VacIPD) | objects.store(Vaccination, NextVT, OptimalSequence, OptimalVacc, VacCar, VacIPD) | ||
cat(" | cat("Funktiot Vaccination, NextVT, OptimalSequence, OptimalVacc sekä ovariablet VacCar, VacIPD tallennettu. \n") | ||
</rcode> | </rcode> | ||
Revision as of 13:55, 4 July 2014
| Moderator:Nobody (see all) Click here to sign up. |
|
|
| Upload data
|
In Opasnet many pages being worked on and are in different classes of progression. Thus the information on those pages should be regarded with consideration. The progression class of this page has been assessed:
|
This page needs a curator. Learn more about curating Opasnet pages. |
Question
How to predict the net effectiveness of a pneumococcal conjugate vaccination with a given set of serotypes when the vaccine is included in the national immunisation programme?
- Focus is on the number of invasive pneumococcal disease (IPD) cases in different age groups.
- The model is assumed to be valid in a population in which an infant pneumococcal conjugate vaccination has been in use for several years s.t. a new steady-state after vaccination has been reached. Coverage of vaccination and vaccine efficacy against carriage are assumed to be high enough to justify the assumtion of full elimination of vaccine type carriage among both the vaccinated and also, due to substancial herd effects, among the unvaccinated members of the population.
- Vaccine type carriage is fully replaced by carriage of the non-vaccine types and the disease causing potential of different serotypes is not altered by vaccination.
Answer
Predicted number of invasive pneumococcal disease (IPD) cases in different age groups are obtained from the serotype replacement model (Nurhonen and Auranen, 2014).
Rationale
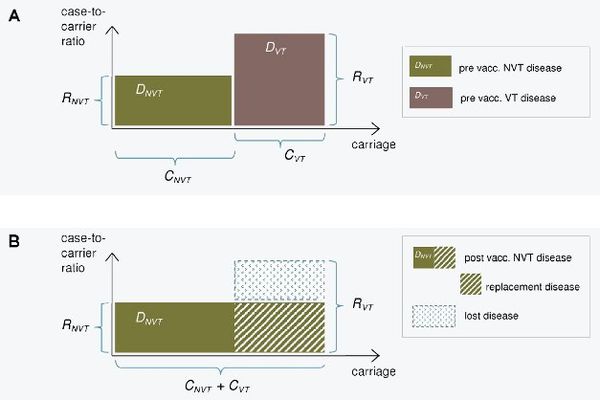
The epidemiological model for pneumococcal carriage and disease is based on the assumption that vaccination completely eliminates the vaccine type carriage in a vaccinated population and this carriage is replaced by non-vaccine type carriage. The implications of this replacement on the decrease or increse in pneumococcal disease then depend on the disease causing potential of the replacing types compared to that of the replaced types. To predict post vaccination disease only pre vaccination data on serotype specific carriage and disease is used.
The consequences of serotype replacement in the model depend on two key assumptions regarding the new steady-state after vaccination:
- the relative serotype proportions among the non-vaccine types are not affected by vaccination (proportionality assumption);
- the case-to-carrier ratios (the disease causing potentials) of individual serotypes remain at their pre-vaccination levels.
The implications of vaccination on disease incidence are assumed to be solely due to the elimination of vaccine type carriage and its replacement by non vaccine type carriage. An exception to this is when a possibility of efficacy against disease without any efficacy against carriage is assumed for certain serotypes.
Computation
The following program illustrates the working of the replacement model. In its current implementation the code allows the user to specify 4 vaccine compositions and then displays the predicted IPD cases in Finland per year corresponding to these vaccines. The results are shown by serotype and by age category (<5 and 5+ year olds). Possible choices for vaccine compositions are: PCV10, PCV13, no vaccination and user specified serotype composition. The program is based on the code in File S1 of Nurhonen and Auranen, 2014.
Instructions for user: Choose the desired vaccine compositions from the list below and then press "Run code". The results will be displayed on a separate tab. The default choices are PCV10 and PCV13.
Funktioiden alustus
See also