Goherr: Fish consumption study: Difference between revisions
(added references) |
|||
| (27 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
{{study|moderator=Arja | {{progression class | ||
|progression=Reviewed | |||
|curator=THL | |||
|date=2019-08-26 | |||
}} | |||
{{study|moderator=Arja}} | |||
:''This page contains a detailed description about data management and analysis of an international survey related to scientific article ''Forage Fish as Food: Consumer Perceptions on Baltic Herring'' by Mia Pihlajamäki, Arja Asikainen, Suvi Ignatius, Päivi Haapasaari, and Jouni T. Tuomisto.<ref name="pihlajamaki2019">Mia Pihlajamäki, Arja Asikainen, Suvi Ignatius, Päivi Haapasaari and Jouni T. Tuomisto. Forage Fish as Food: Consumer Perceptions on Baltic Herring. Sustainability 2019, 11(16), 4298; https://doi.org/10.3390/su11164298</ref> The results of this survey where also used in another article ''Health effects of nutrients and environmental pollutants in Baltic herring and salmon: a quantitative benefit-risk assessment'' by the same group.<ref name="tuomisto2020">Tuomisto, J.T., Asikainen, A., Meriläinen, P., Haapasaari, P. Health effects of nutrients and environmental pollutants in Baltic herring and salmon: a quantitative benefit-risk assessment. BMC Public Health 20, 64 (2020). https://doi.org/10.1186/s12889-019-8094-1</ref> | |||
== Question == | == Question == | ||
| Line 6: | Line 13: | ||
== Answer == | == Answer == | ||
Original questionnaire analysis results | * Model run with all the results of the article<ref name="pihlajamaki2019"/> [http://en.opasnet.org/en-opwiki/index.php?title=Special:RTools&id=B2jMOHfuUSmTjfrn 26.8.2018] | ||
* Original questionnaire analysis results [http://en.opasnet.org/en-opwiki/index.php?title=Special:RTools&id=QaMJZqUX0cPaTfOF 13.3.2017] | |||
* Consumption amount estimates | |||
Consumption amount estimates | ** Model run 21.4.2017 [http://en.opasnet.org/en-opwiki/index.php?title=Special:RTools&id=xc0kaCs8cgzpjwo9] first distribution | ||
* Model run 21.4.2017 [http://en.opasnet.org/en-opwiki/index.php?title=Special:RTools&id=xc0kaCs8cgzpjwo9] first distribution | ** Model run 18.5.2017 [http://en.opasnet.org/en-opwiki/index.php?title=Special:RTools&id=YnuQMDJTQgW1Se5a with modelled data]; [http://en.opasnet.org/en-opwiki/index.php?title=Special:RTools&id=KXXFiP0aj0DYEPdx with direct survey data] | ||
* Model run 18.5.2017 [http://en.opasnet.org/en-opwiki/index.php?title=Special:RTools&id=YnuQMDJTQgW1Se5a with modelled data]; [http://en.opasnet.org/en-opwiki/index.php?title=Special:RTools&id=KXXFiP0aj0DYEPdx with direct survey data] | |||
== Rationale == | == Rationale == | ||
| Line 288: | Line 247: | ||
This code is used to preprocess the original questionnaire data from the above .csv file and to store the data as a usable variable to Opasnet base. The code stores a data.frame named survey. | This code is used to preprocess the original questionnaire data from the above .csv file and to store the data as a usable variable to Opasnet base. The code stores a data.frame named survey. | ||
* Model run 11.7.2018 [http://en.opasnet.org/en-opwiki/index.php?title=Special:RTools&id=Fa8TMg6h8DbEmhjp] | * Model run 11.7.2018 [http://en.opasnet.org/en-opwiki/index.php?title=Special:RTools&id=Fa8TMg6h8DbEmhjp] | ||
* Model run 27.3.2019 with country codes DK EE FI SE [http://en.opasnet.org/en-opwiki/index.php?title=Special:RTools&id=6UJy6JKNE3EyNHEQ] | |||
<rcode name="preprocess2" label="Preprocess (only for developers)"> | <rcode name="preprocess2" label="Preprocess (only for developers)"> | ||
| Line 300: | Line 260: | ||
# Get the data either from Opasnet or your own hard drive. | # Get the data either from Opasnet or your own hard drive. | ||
#survey1 original | #survey1 original fi_le: N:/Ymal/Projects/Goherr/WP5/Goherr_fish_consumption.csv | ||
survey1 <- opasnet.csv( | survey1 <- opasnet.csv( | ||
| Line 306: | Line 266: | ||
wiki = "opasnet_en", sep = ";", fill = TRUE, quote = "\"" | wiki = "opasnet_en", sep = ";", fill = TRUE, quote = "\"" | ||
) | ) | ||
#survey1 <- re#ad.csv( | #survey1 <- re#ad.csv(fi_le = "N:/Ymal/Projects/Goherr/WP5/Goherr_fish_consumption.csv", | ||
# header=FALSE, sep=";", fill = TRUE, quote="\"") | # header=FALSE, sep=";", fill = TRUE, quote="\"") | ||
# Data | # Data fi_le is converted to data.frame using levels at row 2121. | ||
survey1 <- webropol.convert(survey1, 2121, textmark = ":Other open") | survey1 <- webropol.convert(survey1, 2121, textmark = ":Other open") | ||
| Line 329: | Line 289: | ||
ifelse(as.numeric(as.character(survey1$Age)) < 46, "18-45",">45"), | ifelse(as.numeric(as.character(survey1$Age)) < 46, "18-45",">45"), | ||
levels = c("18-45", ">45"), ordered = TRUE | levels = c("18-45", ">45"), ordered = TRUE | ||
) | |||
survey1$Country <- factor( | |||
survey1$Country, | |||
levels=c("DK","EST","FI","SWE"), | |||
labels=c("DK","EE","FI","SE") | |||
) | ) | ||
# Anonymize data | # Anonymize data | ||
| Line 450: | Line 415: | ||
* Sketches about modelling determinants of eating (spring 2018) [http://en.opasnet.org/en-opwiki/index.php?title=Benefit-risk_assessment_of_Baltic_herring_and_salmon_intake&oldid=41947#Sketches_about_modelling_determinants_of_eating] | * Sketches about modelling determinants of eating (spring 2018) [http://en.opasnet.org/en-opwiki/index.php?title=Benefit-risk_assessment_of_Baltic_herring_and_salmon_intake&oldid=41947#Sketches_about_modelling_determinants_of_eating] | ||
==== Figures for the first manuscript ==== | ==== Figures, tables and stat analyses for the first manuscript ==== | ||
* Model run 8.5.2019 with thlVerse code (not run on Opasnet) [http://en.opasnet.org/en-opwiki/index.php?title=Special:RTools&id=46qKuh08I7PHqXRX] [http://en.opasnet.org/en-opwiki/index.php?title=Special:RTools&id=lnq3yNAPXAJFxwaG] | |||
* Model run 26.8.2018 with fig 6 as table [http://en.opasnet.org/en-opwiki/index.php?title=Special:RTools&id=B2jMOHfuUSmTjfrn] | |||
* Previous model runs are [http://en.opasnet.org/en-opwiki/index.php?title=Goherr:_Fish_consumption_study&oldid=42828#Figures.2C_tables_and_stat_analyses_for_the_first_manuscript archived]. | |||
* | Methodological concerns: | ||
* | * [https://stackoverflow.com/questions/12953045/warning-non-integer-successes-in-a-binomial-glm-survey-packages Is the warning in logistic regression important?] | ||
* | * [https://stats.stackexchange.com/questions/46345/how-to-calculate-goodness-of-fit-in-glm-r Goodness of fit in logistic regression] | ||
* | * [https://stats.stackexchange.com/questions/8661/logistic-regression-in-r-odds-ratio Calculate odds ratios in logistic regression] | ||
* [https://www3.nd.edu/~rwilliam/stats3/ordinalindependent.pdf You might treat independent ordinal variables as continuous] | |||
* [https://www.color-blindness.com/coblis-color-blindness-simulator/ Color blindness simulator to adjust colors for the color blind and for black and white printing] | |||
<rcode graphics=1> | <rcode graphics=1> | ||
| Line 463: | Line 434: | ||
library(ggplot2) | library(ggplot2) | ||
library(reshape2) | library(reshape2) | ||
library(MASS) | |||
#library(extrafont) # Needed to save Arial fonts to PDF or EPS | |||
#library(thlVerse) | |||
#library(car) | #library(car) | ||
#library(vegan) | #library(vegan) | ||
BS <- 24 | BS <- 24 | ||
localcomp <- FALSE | |||
thl <- FALSE | |||
groups <- function(o) { | # Set colours | ||
#library(thlVerse) | |||
#library(tidyverse) | |||
#colors <- c( | |||
# thlColors(6,"quali","bar",thin=0.8), | |||
# thlColors(6,"quali","bar",thin=1) | |||
#)[c(1,8,3,10,11,6)] | |||
#ggplot(tibble(A=as.character(1:6),B=1),aes(x=A,weight=B,fill=A))+ | |||
# geom_bar()+ | |||
# scale_fill_manual(values=colors) | |||
colors <- c("#74AF59FF","#2F62ADFF","#D692BDFF","#29A0C1FF","#BE3F72FF","#FBB848FF") | |||
########################## Functions | |||
#### thlLinePlot was adjusted to enable different point shapes | |||
thlLinePlot <- function (data, xvar, yvar, groupvar = NULL, ylabel = yvar, | |||
xlabel = NULL, colors = thlColors(n = 12, type = "quali", | |||
name = "line"), title = NULL, subtitle = NULL, caption = NULL, | |||
legend.position = "none", base.size = 16, linewidth = 3, | |||
show.grid.x = FALSE, show.grid.y = TRUE, lang = "fi", ylimits = NULL, | |||
marked.treshold = 10, plot.missing = FALSE, xaxis.breaks = waiver(), | |||
yaxis.breaks = waiver(), panels = FALSE, nrow.panels = 1, | |||
labels.end = FALSE) | |||
{ | |||
lwd <- thlPtsConvert(linewidth) | |||
gg <- ggplot(data, aes_(x = substitute(xvar), y = substitute(yvar), | |||
group = ifelse(!is.null(substitute(groupvar)), substitute(groupvar), | |||
NA), colour = ifelse(!is.null(substitute(groupvar)), | |||
substitute(groupvar), ""))) + geom_line(size = lwd) | |||
if (isTRUE(plot.missing)) { | |||
df <- thlNaLines(data = data, xvar = deparse(substitute(xvar)), | |||
yvar = deparse(substitute(yvar)), groupvar = unlist(ifelse(deparse(substitute(groupvar)) != | |||
"NULL", deparse(substitute(groupvar)), list(NULL)))) | |||
if (!is.null(df)) { | |||
gg <- gg + geom_line(data = df, aes_(x = substitute(xvar), | |||
y = substitute(yvar), group = ifelse(!is.null(substitute(groupvar)), | |||
substitute(groupvar), NA), colour = ifelse(!is.null(substitute(groupvar)), | |||
substitute(groupvar), "")), linetype = 2, | |||
size = lwd) | |||
} | |||
} | |||
if (!is.null(marked.treshold)) { | |||
if (length(unique(data[, deparse(substitute(xvar))])) > | |||
marked.treshold) { | |||
if (is.factor(data[, deparse(substitute(xvar))]) || | |||
is.character(data[, deparse(substitute(xvar))]) || | |||
is.logical(data[, deparse(substitute(xvar))])) { | |||
levs <- levels(factor(data[, deparse(substitute(xvar))])) | |||
min <- levs[1] | |||
max <- levs[length(levs)] | |||
} | |||
else { | |||
min <- min(data[, deparse(substitute(xvar))]) | |||
max <- max(data[, deparse(substitute(xvar))]) | |||
} | |||
subdata <- data[c(data[, deparse(substitute(xvar))] %in% | |||
c(min, max)), ] | |||
gg <- gg + geom_point(data = subdata, aes_( | |||
x = substitute(xvar), | |||
y = substitute(yvar), | |||
group = ifelse(!is.null(substitute(groupvar)), substitute(groupvar), NA), | |||
colour = ifelse(!is.null(substitute(groupvar)), substitute(groupvar), ""), | |||
shape = substitute(groupvar)), | |||
fill = "white", | |||
stroke = 1.35 * lwd, | |||
size = 10/3 * lwd)+scale_shape_manual(values=21:25) | |||
} | |||
else { | |||
gg <- gg + geom_point( | |||
aes_( | |||
shape = substitute(groupvar) | |||
), | |||
fill = "white", | |||
stroke = 1.35 * lwd, | |||
size = 10/3 * lwd)+scale_shape_manual(values=21:25) | |||
} | |||
} | |||
if (isTRUE(labels.end)) { | |||
if (is.factor(data[, deparse(substitute(xvar))]) || | |||
is.character(data[, deparse(substitute(xvar))]) || | |||
is.logical(data[, deparse(substitute(xvar))])) { | |||
levs <- levels(factor(data[, deparse(substitute(xvar))])) | |||
maxd <- data[data[, deparse(substitute(xvar))] == | |||
levs[length(levs)], ] | |||
} | |||
else { | |||
maxd <- data[data[, deparse(substitute(xvar))] == | |||
max(data[, deparse(substitute(xvar))]), ] | |||
} | |||
brks <- maxd[, deparse(substitute(yvar))] | |||
labsut <- maxd[, deparse(substitute(groupvar))] | |||
} | |||
else (brks <- labsut <- waiver()) | |||
gg <- gg + ylab(ifelse(deparse(substitute(ylabel)) == "yvar", | |||
deparse(substitute(yvar)), ylabel)) + labs(title = title, | |||
subtitle = subtitle, caption = caption) + thlTheme(show.grid.y = show.grid.y, | |||
show.grid.x = show.grid.x, base.size = base.size, legend.position = legend.position, | |||
x.axis.title = ifelse(!is.null(xlabel), TRUE, FALSE)) + | |||
xlab(ifelse(!is.null(xlabel), xlabel, "")) + scale_color_manual(values = colors) + | |||
thlYaxisControl(lang = lang, limits = ylimits, breaks = yaxis.breaks, | |||
sec.axis = labels.end, sec.axis.breaks = brks, sec.axis.labels = labsut) | |||
if (is.factor(data[, deparse(substitute(xvar))]) || is.character(data[, | |||
deparse(substitute(xvar))]) || is.logical(data[, deparse(substitute(xvar))])) { | |||
gg <- gg + scale_x_discrete(breaks = xaxis.breaks, expand = expand_scale(mult = c(0.05))) | |||
} | |||
else (gg <- gg + scale_x_continuous(breaks = xaxis.breaks)) | |||
if (isTRUE(panels)) { | |||
fmla <- as.formula(paste0("~", substitute(groupvar))) | |||
gg <- gg + facet_wrap(fmla, scales = "free", nrow = nrow.panels) | |||
} | |||
gg | |||
} | |||
groups <- function(o) { | |||
o$Group <- paste(o$Gender, o$Ages) | o$Group <- paste(o$Gender, o$Ages) | ||
o$Group <- factor(o$Group, levels = c( | o$Group <- factor(o$Group, levels = c( | ||
| Line 478: | Line 570: | ||
return(o) | return(o) | ||
} | } | ||
#' @title reasons produces a ggplot object for a graph that shows fractions of reasons mentioned in the data. | |||
#' @param dat a data.frame containing survey answers for the target subgroup. | |||
#' @param title an atomic string with the name to be shown as the title of graph | |||
#' @return ggplot object | |||
reasons <- function( | reasons <- function( | ||
dat, | dat, | ||
title | title | ||
) { | ) { | ||
require(reshape2) | |||
weight <- Ovariable("weight",data=data.frame( | weight <- Ovariable("weight",data=data.frame( | ||
Row=dat$Row, | Row=dat$Row, | ||
| Line 499: | Line 597: | ||
tmp$Result <- (tmp$Result %in% c("Yes","1"))*1 | tmp$Result <- (tmp$Result %in% c("Yes","1"))*1 | ||
tmp <- weight * (Ovariable("tmp",data=tmp)) | tmp <- weight * (Ovariable("tmp",data=tmp)) | ||
cat(title, ": number of individual answers.\n") | |||
oprint(oapply(tmp, c("tmpResult","Country"),length)@output) | oprint(oapply(tmp, c("tmpResult","Country"),length)@output) | ||
| Line 506: | Line 605: | ||
tmp$Reason <- factor(tmp$Reason, levels=popu) | tmp$Reason <- factor(tmp$Reason, levels=popu) | ||
levels(tmp$Reason) <- gsub("( BH| BS)", "", gsub("\\.", " ", levels(tmp$Reason))) | levels(tmp$Reason) <- gsub("( BH| BS)", "", gsub("\\.", " ", levels(tmp$Reason))) | ||
cat(title, ": fractions shown on graph.\n") | |||
oprint(tmp@output) | |||
tmp <- ggplot(tmp@output, aes(x=Reason, y=Result,colour=Country, group=Country))+ | if(thl) { | ||
tmp <- thlLinePlot(tmp@output, xvar=Reason, yvar=Result,groupvar=Country, | |||
colors= c("#519B2FFF", "#2F62ADFF", "#BE3F72FF","#88D0E6FF"), # #29A0C1FF"), | |||
# THL colors but fourth is brigter | |||
legend.position = c(0.85,0.2), base.size = BS, title=title, | |||
subtitle="Fraction of population")+ | |||
coord_flip()+ | |||
scale_y_continuous(labels=scales::percent_format(accuracy=1)) | |||
} else { | |||
tmp <- ggplot(tmp@output, aes(x=Reason, y=Result,colour=Country, group=Country))+ | |||
geom_point(shape=21, size=5, fill="Grey", stroke=2)+ | |||
geom_line(size=1.2)+ | |||
coord_flip()+ | |||
theme_gray(base_size=BS)+ | |||
scale_y_continuous(labels=scales::percent_format())+#accuracy=1))+ | |||
scale_colour_manual(values=colors)+ | |||
labs( | |||
title=title, | |||
x="Answer", | |||
y="Fraction of population") | |||
} | |||
return(tmp) | return(tmp) | ||
} | } | ||
impacts <- function(dat,title) { | #' @title Function impacts produces a graph showing the magnitude of impact if some things change | ||
#' @param dat data.frame containing answers | |||
#' @param title string to be used as the title of the graph | |||
#' @param population ovariable of country population weights | |||
#' @return ggplot object | |||
impacts <- function(dat,title, population) { | |||
tmp <- melt( | tmp <- melt( | ||
dat, | dat, | ||
id.vars = "Country", | id.vars = c("Country","Weighting"), | ||
variable.name ="Reason", | variable.name ="Reason", | ||
value.name="Response" | value.name="Response" | ||
| Line 552: | Line 670: | ||
)] <- "Increase" | )] <- "Increase" | ||
popu <- aggregate(as.numeric(tmp$Response), tmp["Reason"],sum) | colnames(tmp)[colnames(tmp)=="Weighting"] <- "Result" | ||
tmp <- Ovariable("tmp",data=tmp) * population | |||
popu <- aggregate(as.numeric(tmp$Response), tmp@output["Reason"],sum) | |||
popu <- popu$Reason[order(popu$x)] | popu <- popu$Reason[order(popu$x)] | ||
tmp$Reason <- factor(tmp$Reason, levels=popu) | tmp$Reason <- factor(tmp$Reason, levels=popu) | ||
levels(tmp$Reason) <- gsub("( BH| BS)", "", gsub("\\.", " ", levels(tmp$Reason))) | levels(tmp$Reason) <- gsub("( BH| BS)", "", gsub("\\.", " ", levels(tmp$Reason))) | ||
tmp <- (oapply(tmp, c("Reason","Response"), sum)/ | |||
oapply(tmp, c("Reason"),sum)) | |||
tmp <- (oapply(tmp, c( | |||
oapply(tmp, c( | |||
oprint(tmp@output) | |||
print(ggplot(tmp@output, | |||
aes(x=Reason, weight=Result, fill=Response))+geom_bar()+ | |||
coord_flip()+#facet_grid(.~Country)+ | |||
theme_gray(base_size=BS)+ | |||
theme(legend.position = "bottom")+ | |||
scale_fill_manual(values=c("Grey",colors[c(1,6,5)]))+ | |||
scale_y_continuous(breaks=c(0.5,1),labels=scales::percent_format())+ | |||
labs( | |||
title=title, | |||
x="Cause", | |||
y="Fraction of population" | |||
)) | |||
return(tmp@output) | |||
} | |||
#' @title or95 prints the odds ratio and 95 % confidence interval of a glm fit | |||
#' @param fit a glm fit | |||
#' @return data.frame with OR and 95%CI | |||
or95 <- function(fit) { | |||
cat("Odds ratios with 95 % confidence intervals\n") | |||
tmp <- exp(cbind(coef(fit), confint(fit))) | |||
tmp <- data.frame( | |||
rownames(tmp), | |||
paste0(sprintf("%.2f",tmp[,1]), " (",sprintf("%.2f", tmp[,2]),"-",sprintf("%.2f",tmp[,3]),")") | |||
) | |||
colnames(tmp) <- c("Parameter","OR (95% CI)") | |||
return(tmp) | return(tmp) | ||
} | } | ||
##### | #' @title Perform and print regression analyses | ||
#' @param dat data.frame with data | |||
#' @param y name of column to be used as dependent variable | |||
#' @param countries logical atom to tell whether to run country-specific analyses | |||
#' @return returns the model fit object | |||
printregr <- function(dat, y, title, countries = TRUE) { | |||
dat$y <- dat[[y]] | |||
cat("\n############### Logistic regression analysis:",title, "(", y, ")\n") | |||
fit <- glm( | |||
y ~ Ages + Gender + Country + as.numeric(Education) + as.numeric(Purchasing.power), | |||
family=binomial(),na.action="na.omit",weights=Weighting, | |||
data=dat | |||
) | |||
step <- stepAIC(fit, direction = "both") | |||
oprint(step$anova) | |||
oprint(summary(fit)) | |||
oprint(or95(fit)) | |||
if(countries) { | |||
for(i in unique(dat$Country)) { | |||
cat("\n#### Country-specific logistic regression analysis:", i, y, "\n") | |||
fit <- glm( | |||
Eat.fish ~ Ages + Gender + as.numeric(Education) + as.numeric(Purchasing.power), | |||
family=binomial(),na.action="na.omit",weights=Weighting, | |||
data=dat[dat$Country==i,] | |||
) | |||
step <- stepAIC(fit, direction = "both") | |||
oprint(step$anova) | |||
oprint(summary(fit)) | |||
oprint(or95(fit)) | |||
} | |||
} | |||
return(fit) | |||
} | |||
#### | #################### Data for Figure 1. | ||
su1 <- EvalOutput(Ovariable( | if(thl) { # Commented out because needs package thlVerse. | ||
"su1", | |||
data = data.frame( | dat <- opbase.data("Op_en7749", subset="Fish consumption as food in Finland") # [[Goherr: Fish consumption study]] | ||
survey1[c("Country","Gender","Education","Purchasing.power")], | dat$Year <- as.numeric(as.character(dat$Year)) | ||
Result=1 | |||
gr <- thlLinePlot( | |||
dat[dat$Species=="Total",], | |||
xvar=Year, | |||
yvar=Result, | |||
ylimits=c(0,12), | |||
groupvar=Origin, | |||
base.size=24, | |||
title="Fish consumption in Finland", | |||
ylabel="", | |||
subtitle="(kg/a per person)", | |||
legend.position = "bottom" | |||
) | |||
if(localcomp) ggsave("Figure 1.pdf", width=8,height=6) | |||
if(localcomp) ggsave("Figure 1.png", width=8,height=6) | |||
} # End if | |||
################## Get data | |||
objects.latest("Op_en7749", "preprocess2") # [[Goherr: Fish consumption study]]: survey1, surv | |||
objects.latest("Op_en7749", "nonsamplejsp") # [[Goherr: Fish consumption study]]: jsp every respondent from data exactly once. | |||
objects.latest("Op_en7749", "initiate") # [[Goherr: Fish consumption study]]: amount etc. | |||
openv.setN(50) | |||
survey1 <- groups(survey1) | |||
#levels(survey1$Country) <- c("FI","SE","DK","EE") # WHY was this here? It is a potential hazard. | |||
effinfo <- 0 # No policies implemented. | |||
effrecomm <- 0 | |||
population <- EvalOutput(Ovariable( | |||
"population", | |||
ddata = "Op_en7748", | |||
subset = "Population" | |||
)) | |||
levels(population$Country) <- c("DK","EE","FI","SE") | |||
population <- oapply(population, c("Country"),sum) | |||
amount <- EvalOutput(amount) | |||
#result(amount)[result(amount)==0] <- 0.1 Not needed if not log-transformed. | |||
amount <- amount * 365.25 / 1000 # g/day --> kg/year | |||
#### General fish eating | |||
su1 <- EvalOutput(Ovariable( | |||
"su1", | |||
data = data.frame( | |||
survey1[c("Country","Gender","Education","Purchasing.power")], | |||
Result=1 | |||
) | ) | ||
)) | )) | ||
# Figure 1. Origin of consumed fish in Finland between 1999 and 2016. | |||
# Data not on this page, drawn separately. | |||
# Table 1. Dimensions of embeddedness, modified from Hass (2007, p. 16) | |||
# Written directly on the manuscript. | |||
# | # Table 2: statistics of the survey population in each country (n, female %, education, purchasing power) | ||
tmp <- round(t(data.frame( | |||
survey1 | n = tapply(rep(1,nrow(survey1)), survey1$Country,sum), | ||
" | Females = tapply(survey1$Gender=="Female", survey1$Country,mean)*100, | ||
) | Old = tapply(survey1$Ages==">45", survey1$Country,mean)*100, | ||
PurcVlow = tapply(survey1$Purchasing.power=="Very low", survey1$Country,mean)*100, | |||
PurcLow = tapply(survey1$Purchasing.power=="Low", survey1$Country,mean)*100, | |||
" | PurcSuf = tapply(survey1$Purchasing.power=="Sufficient", survey1$Country,mean)*100, | ||
) | PurcGood = tapply(survey1$Purchasing.power=="Good", survey1$Country,mean)*100, | ||
PurcVgood = tapply(survey1$Purchasing.power=="Very good", survey1$Country,mean)*100, | |||
PurcExc = tapply(survey1$Purchasing.power=="Excellent", survey1$Country,mean)*100, | |||
" | EducPri = tapply(survey1$Education=="Primary education", survey1$Country,mean)*100, | ||
) | EducSec = tapply(survey1$Education=="Secondary education (gymnasium, vocational school or similar)", survey1$Country,mean)*100, | ||
EducCol = tapply(survey1$Education=="Lower level college education or similar", survey1$Country,mean)*100, | |||
EducHig = tapply(survey1$Education=="Higher level college education or similar", survey1$Country,mean)*100 | |||
" | ))) | ||
rownames(tmp) <- c( | |||
"Number of respondents", | |||
"Females (%)", | |||
">45 years (%)", | |||
"Purchasing power: Very low (%)", | |||
"Low (%)", | |||
"Sufficient (%)", | |||
"Good (%)", | |||
"Very good (%)", | |||
"Excellent (%)", | |||
"Education: Primary education (%)", | |||
"Secondary education (%)", | |||
"Lower level college (%)", | |||
"Higher level college (%)" | |||
) | ) | ||
oprint(tmp, digits=0) | |||
cat("Weighted fraction of fish eaters\n") | |||
tmp <- aggregate(survey1$Weighting, survey1[c("Eat.fish","Country")],FUN=sum) | |||
oprint(aggregate(tmp$x, tmp["Country"],FUN=function(x) x[2]/sum(x))) | |||
) | |||
ggplot(survey1, | ggplot(survey1, | ||
aes(x= | aes(x=Eat.fish, weight=Weighting, group=Country), stat="count")+ | ||
geom_bar(aes(y=..prop.., fill=Country), position="dodge")+ | |||
theme_gray(base_size=BS)+ | theme_gray(base_size=BS)+ | ||
scale_fill_manual(values=colors)+ | |||
scale_y_continuous(labels=scales::percent_format())+ | scale_y_continuous(labels=scales::percent_format())+ | ||
labs(title="Fraction of fish eaters within population") | |||
labs(title=" | |||
############## Answers to open questions | |||
if(FALSE){ | |||
cat("Number of open-ended answers per question and country\n") | |||
tmp <- survey1[c(1,3,27:28,36:37,44:45,60:61,71:72,83:84,93:94,109:110,121:122,133:134,154,157:158)] | |||
tmp <- reshape( | |||
tmp, | |||
# | varying=list( | ||
Condition = colnames(tmp)[grepl("other",tolower(colnames(tmp)))], | |||
Text = colnames(tmp)[grepl("open",colnames(tmp))] | |||
), | |||
times = colnames(tmp)[grepl("other",tolower(colnames(tmp)))], | |||
direction="long" | |||
) | |||
tmp <- tmp[tmp$Why.not.fish.open!="" , ] | |||
oprint(table(tmp$time,tmp$Country)) | |||
oprint(tmp) | |||
} # End if(FALSE) | |||
#### Baltic salmon | |||
reasons( | reasons( | ||
survey1[survey1$Eat. | survey1[survey1$Eat.fish=="No"& !is.na(survey1$Eat.fish),c(1,17:26,154,157)], # 1 Country, 8 Why don't you eat fish | ||
"Reasons not to eat among fish non-consumers" | |||
"Reasons not to eat among | |||
) | ) | ||
# Figure 5 | |||
cat("Figure 5 (to be converted to text)\n") | |||
reasons( | |||
survey1[survey1$Eat.salmon=="Yes"& !is.na(survey1$Eat.salmon),c(1,31:36,154,157)], # 1 Country, 11 Which salmon species | |||
"Species used among salmon consumers" | |||
) | |||
reasons( | |||
survey1[survey1$Baltic.salmon=="Yes"& !is.na(survey1$Baltic.salmon),c(1,50:59,154,157)], # 1 Country, 17 Why Baltic salmon | |||
"Reasons to eat among Baltic salmon consumers" | |||
) | |||
=== | if(localcomp) ggsave("Figure 5.pdf", width=10,height=5) | ||
if(localcomp) ggsave("Figure 5.png", width=10,height=5) | |||
reasons( | |||
survey1[survey1$Baltic.salmon=="No"& !is.na(survey1$Baltic.salmon),c(1,62:70,154,157)], # 1 Country, 18 Why not Baltic salmon | |||
"Reasons not to eat, Baltic salmon non-consumers" | |||
) | |||
if(localcomp) ggsave("Figure 6.pdf", width=10,height=5) | |||
if(localcomp) ggsave("Figure 6.png", width=10,height=5) | |||
impacts.sal <- impacts( | |||
survey1[survey1$Baltic.salmon=="Yes"& !is.na(survey1$Baltic.salmon),c(1,73:82,154)], | |||
"Effect on Baltic salmon consumption", | |||
population | |||
) | |||
####### Any herring | |||
ggplot(survey1[survey1$Eat.fish=="Yes",], | |||
aes(x=Country, weight=Weighting, fill=Eat.herring))+geom_bar(position="fill")+ | |||
coord_flip()+#facet_grid(.~Country)+ | |||
theme_gray(base_size=BS)+ | |||
theme(legend.position = "bottom")+ | |||
scale_y_continuous(labels=scales::percent_format())+ | |||
scale_fill_manual(values=colors)+ | |||
labs( | |||
title="Any herring consumption", | |||
y="Fraction of fish consumers" | |||
) | |||
####### Baltic herring | |||
### Figure 2. Comparison of Baltic herring consumption habits. | |||
survey1$What <- ifelse(is.na(survey1$Eat.BH), "Other fish", as.character(survey1$Eat.BH)) | |||
survey1$What <- factor( | |||
survey1$What, | |||
levels=(c("Yes","I don't know","No","Other fish")), | |||
labels=(c("Baltic herring","Some herring","Other herring","Other fish")) | |||
) | |||
tmp <- survey1[survey1$Eat.fish=="Yes",] | |||
colnames(tmp)[colnames(tmp)=="Weighting"] <- "Result" | |||
tmp <- EvalOutput(Ovariable("tmp", data=tmp)) | |||
tmp <- oapply(tmp, c("What","Country"), sum) / oapply(tmp,"Country",sum) | |||
tmp$Country <- factor(tmp$Country, levels=rev(levels(tmp$Country))) | |||
if(thl) { | |||
thlBarPlot(tmp@output, xvar=Country, yvar=Result, groupvar=What, horizontal = TRUE, stacked = TRUE, | |||
legend.position = "bottom",, | |||
title="Baltic herring consumption", | |||
subtitle="Fraction of fish consumers", | |||
)+ | |||
scale_y_continuous(labels=scales::percent_format()) | |||
} else { | |||
ggplot(tmp@output, aes(x=Country, weight=Result, fill=What))+geom_bar(position="fill")+ | |||
coord_flip()+#facet_grid(.~Country)+ | |||
theme_gray(base_size=BS)+ | |||
theme(legend.position = "bottom")+ | |||
scale_y_continuous(labels=scales::percent_format())+ | |||
scale_fill_manual(values=c("gray",colors))+ | |||
guides(fill=guide_legend(reverse=TRUE))+ | |||
labs( | |||
title="Baltic herring consumption", | |||
y="Fraction of fish consumers" | |||
) | ) | ||
} | |||
if(localcomp) ggsave("Figure 2.pdf",width=10, height=5) | |||
if(localcomp) ggsave("Figure 2.png",width=6, height=3) | |||
tmp <- survey1[survey1$Eat.fish=="Yes",] | |||
tmp <- aggregate(tmp$Weighting, tmp[c("What","Country")], sum) | |||
rown <- tmp$What[tmp$Country=="FI"] | |||
tmp <- aggregate(tmp$x, tmp["Country"],FUN=function(x) x/sum(x)) | |||
colnames(tmp[[2]]) <- rown | |||
oprint(as.matrix(tmp)) | |||
ggplot(survey1[survey1$Eat.herring=="Yes",], | |||
aes(x=Country, weight=Weighting, fill=Eat.BH))+geom_bar(position="fill")+ | |||
coord_flip()+#facet_grid(.~Country)+ | |||
theme_gray(base_size=BS)+ | |||
theme(legend.position = "bottom")+ | |||
scale_y_continuous(labels=scales::percent_format())+ | |||
scale_fill_manual(values=c(colors))+ | |||
labs( | |||
title="Baltic herring consumption", | |||
y="Fraction of herring consumers" | |||
) | |||
####### | ####### Any salmon | ||
ggplot(survey1[survey1$Eat.fish=="Yes",], | |||
aes(x=Country, weight=Weighting, fill=Eat.salmon))+geom_bar(position="fill")+ | |||
coord_flip()+ | |||
theme_gray(base_size=BS)+ | |||
theme(legend.position = "bottom")+ | |||
scale_y_continuous(labels=scales::percent_format())+ | |||
scale_fill_manual(values=colors)+ | |||
labs( | |||
title="Any salmon consumption", | |||
y="Fraction of fish consumers" | |||
) | |||
####### Baltic salmon | |||
ggplot(survey1[survey1$Eat.fish=="Yes",], | |||
aes(x=Country, weight=Weighting, fill=Baltic.salmon))+geom_bar(position="fill")+ | |||
coord_flip()+ | |||
theme_gray(base_size=BS)+ | |||
theme(legend.position = "bottom")+ | |||
scale_y_continuous(labels=scales::percent_format())+ | |||
scale_fill_manual(values=colors)+ | |||
labs( | |||
title="Baltic salmon consumption", | |||
y="Fraction of fish consumers" | |||
) | |||
ggplot(survey1[survey1$Eat.salmon=="Yes",], | |||
aes(x=Country, weight=Weighting, fill=Baltic.salmon))+geom_bar(position="fill")+ | |||
coord_flip()+ | |||
theme_gray(base_size=BS)+ | |||
theme(legend.position = "bottom")+ | |||
scale_y_continuous(labels=scales::percent_format())+ | |||
scale_fill_manual(values=colors)+ | |||
labs( | |||
title="Baltic salmon consumption", | |||
y="Fraction of salmon consumers" | |||
) | |||
############## Recommendation awareness | |||
levels(survey1$Recommendation.awareness) <- c("Not aware","Aware","Know content") | |||
ggplot(survey1, | |||
aes(x=Country, weight=Weighting, fill=Recommendation.awareness))+geom_bar(position="fill")+ | |||
coord_flip()+#facet_grid(.~Country)+ | |||
theme_gray(base_size=BS)+ | |||
theme(legend.position = "bottom")+ | |||
scale_y_continuous(labels=scales::percent_format())+ | |||
scale_fill_manual(values=colors)+ | |||
labs(title="Awareness of food recommendations about Baltic fish") | |||
# | # Figure 3. Percentages of reasons to eat Baltic herring | ||
reasons( | |||
survey1[survey1$Eat.BH=="Yes"& !is.na(survey1$Eat.BH),c(1,99:108,154,157)], # 1 Country, 27 why Baltic herring | |||
"Reasons to eat among Baltic herring consumers" | |||
) | |||
if(localcomp) ggsave("Figure 3.pdf", width=10, height=5) | |||
if(localcomp) ggsave("Figure 3.png", width=10, height=5) | |||
# | # Figure 4. Reasons for not to eat Baltic herring. | ||
# Denmark and Estonia were omitted because they had less than 20 observations. | |||
reasons( | |||
survey1[ | |||
survey1$Eat.BH=="No"& | |||
survey1$Country %in% c("FI","SE") & | |||
!is.na(survey1$Eat.BH),c(1,111:120,154,157)], # 1 Country, 28 Why not Baltic herring | |||
"Reasons not to eat, Baltic herring non-consumers" | |||
)+scale_color_manual(values=c("#BE3F72FF","#88D0E6FF"))+ | |||
scale_shape_manual(values=23:24) | |||
if(localcomp) ggsave("Figure 4.pdf", width=10, height=5) | |||
if(localcomp) ggsave("Figure 4.png", width=10, height=5) | |||
# Figure 7. The role of different determinants on Baltic salmon consumption | |||
impacts.herr <- impacts( | |||
survey1[survey1$Eat.BH=="Yes"& !is.na(survey1$Eat.BH),c(1,123:132,154)], # 1 Country, 29 Influence of Baltic herring policies | |||
title="Effect on Baltic herring consumption", | |||
population | |||
) | |||
tmp <- rbind( | |||
data.frame( | |||
Fish="Herring", | |||
impacts.herr | |||
), | |||
data.frame( | |||
Fish="Salmon", | |||
impacts.sal | |||
) | |||
) | |||
oprint(aggregate(tmp$Result,tmp[c("Reason","Response","Fish")],sum)) | |||
if(thl) { | |||
tmp$Response <- factor(tmp$Response, levels=rev(levels(tmp$Response))) | |||
thlBarPlot(aggregate(tmp$Result,tmp[c("Reason","Response","Fish")],sum), | |||
xvar=Reason, yvar=x, groupvar=Response, legend.position = "bottom", | |||
horizontal = TRUE, stacked = TRUE, | |||
base.size = BS, title="Effect on Baltic fish consumption", | |||
subtitle="Fraction of population")+ | |||
facet_grid(.~Fish)+ | |||
scale_fill_manual(values=rev(c("gray",colors[c(1,6,5)])))+ | |||
scale_y_continuous(breaks=c(0.5,1),labels=scales::percent_format(accuracy=1)) | |||
} else { | |||
ggplot(tmp, aes(x=Reason, weight=Result, fill=Response))+geom_bar()+ | |||
coord_flip()+facet_grid(.~Fish)+ | |||
theme_gray(base_size=BS)+ | |||
theme(legend.position = "bottom")+ | |||
scale_fill_manual(values=c("gray",colors[c(1,6,5)]))+ | |||
scale_y_continuous(breaks=c(0.5,1),labels=scales::percent_format())+#accuracy=1))+ | |||
guides(fill=guide_legend(reverse=TRUE))+ | |||
labs( | |||
title="Effect on Baltic fish consumption", | |||
x="Cause", | |||
y="Fraction of population" | |||
) | |||
} | } | ||
if(localcomp) ggsave("Figure 7.pdf", width=12, height=7) | |||
if(localcomp) ggsave("Figure 7.png", width=12, height=7) | |||
######### | ######### Amounts estimated for each respondent | ||
amount <- groups(amount) | |||
#tmp <- amount*info | |||
#tmp <- tmp[ | |||
# tmp$Info.improvements=="BAU" & | |||
# tmp$Recomm.herring=="BAU" & | |||
# tmp$Recomm.salmon=="BAU" , | |||
# ] | |||
tmp <- cbind( | |||
oapply(amount, c("Country","Group","Gender","Ages","Fish"), FUN=mean)@output, | |||
SD=oapply(amount, c("Country","Group","Gender","Ages","Fish"), FUN=sd)$amountResult | |||
) | |||
## | # Figure 3 (old, unused). Average consumption (kg/year) of Baltic herring in four countries, | ||
# calculated with Monte Carlo simulation (1000 iterations). | |||
ggplot(tmp[tmp$Fish=="Herring",], aes(x=Group, weight=amountResult, fill=Group))+ | |||
geom_bar()+facet_wrap(~Country)+ | |||
theme_gray(base_size=BS)+ | |||
scale_fill_manual(values=colors)+ | |||
guides(fill=FALSE)+ | |||
theme(axis.text.x = element_text(angle = -90))+ | |||
labs( | |||
title="Baltic herring consumption in subgroups", | |||
y="Average consumption (kg/year)") | |||
# if(localcomp) ggsave("Figure3.pdf", width=9, height=10) | |||
# if(localcomp) ggsave("Figure3.png", width=9, height=10) | |||
tmp$Result <- paste0(sprintf("%.1f",tmp$amountResult), " (",sprintf("%.1f", tmp$SD,1),")") | |||
tmp <- reshape(tmp, v.names="Result", timevar="Country", idvar=c("Fish","Group"),drop=c("Gender","Ages","amountResult","SD"), direction="wide") | |||
colnames(tmp) <- gsub("Result\\.", "", colnames(tmp)) | |||
cat("Average fish consumption in subgroups, mean (sd)\n") | |||
oprint(tmp) | |||
weight <- EvalOutput(Ovariable("weight",data=data.frame( | |||
amount@output[c("Row","Iter","Fish","Country")], | |||
Result=amount$Weighting | |||
))) | |||
</ | tmp <- (oapply(amount * weight, c("Fish","Country"), sum) / oapply(weight,c("Fish","Country"), sum))@output | ||
cat("Average fish consumption per country (kg/year)\n") | |||
oprint(tmp) | |||
[ | result(amount)[result(amount)==0] <- 0.1 | ||
ggplot(amount@output, aes(x=amountResult, colour=Group))+stat_ecdf()+scale_x_log10()+ | |||
facet_wrap(~Fish) | |||
############################ | |||
#### Statistical analyses | |||
dat <- merge(survey1,amount@output) # [amount$Fish=="Herring",]) # This results in 50 * 2 times of rows | |||
* | |||
# These should be corrected to preprocess2. NOT ordered. | |||
dat$Country <- factor(dat$Country, ordered=FALSE) | |||
dat$Ages <- factor(dat$Ages, ordered=FALSE) | |||
dat$Gender <- factor(dat$Gender, ordered=FALSE) | |||
ggplot(dat, aes(x=amountResult, weight=Weighting, color=Country))+stat_ecdf()+facet_wrap(~Fish)+ | |||
scale_x_log10() | |||
gro <- unique(dat$Group) | |||
out <- list(data.frame(), data.frame()) | |||
for(i in unique(dat$Fish)) { | |||
for(j in unique(dat$Country)){ | |||
< | for(k in unique(dat$Iter)) { | ||
res <- kruskal.test(amountResult ~ Group, data=dat[ | |||
dat$Iter==k & dat$Country==j & dat$Fish==i,]) | |||
out[[1]] <- rbind(out[[1]], data.frame( | |||
test="Kruskal-Wallis", | |||
Fish=i, | |||
Country=j, | |||
Iter=k, | |||
p.value=res[[3]]) | |||
) | |||
for(l in 1:(length(gro)-1)) { | |||
for(m in (l+1):length(gro)) { | |||
res2 <- wilcox.test( | |||
x=dat$amountResult[dat$Iter==k & dat$Country==j & dat$Fish==i & dat$Group==gro[l]], | |||
y=dat$amountResult[dat$Iter==k & dat$Country==j & dat$Fish==i & dat$Group==gro[m]], conf.int = FALSE) | |||
out[[2]] <- rbind(out[[2]], data.frame( | |||
Fish=i, | |||
Country=j, | |||
Iter=k, | |||
Pair1=gro[l], | |||
Pair2=gro[m], | |||
p.value = res2$p.value | |||
)) | |||
} | |||
} | |||
} | |||
} | |||
} | |||
ggplot(out[[1]], aes(x=paste(Fish, Country), y = p.value, colour = Country))+geom_jitter()+ | |||
geom_hline(yintercept=0.05)+coord_flip()+theme_gray(base_size=BS)+ | |||
labs(title="Kruskal-Wallis non-parametric test across all groups") | |||
ggplot(out[[2]], aes(x=paste(Fish, Country), y=p.value, colour=Pair1, shape=Pair2))+geom_jitter()+ | |||
geom_hline(yintercept=0.05)+coord_flip()+theme_gray(base_size=BS)+ | |||
labs(title="Mann-Whitney U test across all pairs of groups") | |||
cat("Kurskal-Wallis non-parametric test for 50 iterations of amount\n") | |||
oprint(aggregate(out[[1]]["p.value"], out[[1]][c("Country","Fish")], mean)) | |||
cat("Mann-Whitney U non-parametric test for 50 iterations of amount\n") | |||
oprint(aggregate(out[[2]]["p.value"], out[[2]][c("Pair1","Pair2","Country","Fish")], mean)) | |||
###################### Logistic regression | |||
dat2 <- dat[dat$Iter==1 & dat$Fish=="Herring",] | |||
tmp <- printregr(dat2, "Eat.fish", "What explains whether people eat fish at all?") | |||
tmp <- printregr(dat2, "Eat.herring", "What explains whether people eat herring compared with other fish?") | |||
dat2$Eat.BH2 <- ifelse(dat2$Eat.BH!="Yes" | is.na(dat2$Eat.BH),0,1) | |||
tmp <- printregr(dat2, "Eat.BH2", "What explains whether people eat Baltic herring compared with everyone else?") | |||
dat2$What2 <- ifelse(dat2$What=="Baltic herring",1,ifelse(dat2$What=="Other herring",0,NA)) | |||
tmp <- printregr(dat2, "What2", "What explains whether people eat Baltic herring compared with other herring?") | |||
cat("###################### How fish-specific causes are explained by | |||
population determinants?\n") | |||
for(j in c( | |||
"Tastes.good.BH", | |||
"Self.caught.BH", | |||
"Easy.to.cook.BH", | |||
"Quick.to.cook.BH", | |||
"Readily.available.BH", | |||
"Healthy.BH", | |||
"Inexpensive.BH", | |||
"Family.likes.it.BH", | |||
"Environmental.BH", | |||
"Traditional.BH" | |||
)) { | |||
tmp <- printregr(dat2, j, "What explains the reason to eat Baltic herring?") | |||
} | |||
for(j in c( | |||
"Bad.taste.BH", | |||
"Not.used.to.BH", | |||
"Cannot.cook.BH", | |||
"Difficult.to.cook.BH", | |||
"Not.available.BH", | |||
"Health.risks.BH", | |||
"Better.for.feed.BH", | |||
"Quality.issues.BH", | |||
"Sustainability.BH", | |||
"Not.traditional.BH" | |||
)) { | |||
tmp <- printregr(dat2, j, "What explains the reason to not eat Baltic herring?") | |||
} | |||
# tmp <- printregr(dat2, "Eat.salmon", "What explains whether people eat any salmon compared with other fish?") | |||
# tmp <- printregr(dat2, "Baltic.salmon", "What explains whether people eat Baltic salmon compared with other salmon?") | |||
</rcode> | |||
Luke data about fish consumption in Finland [https://stat.luke.fi/en/fish-consumption-2017_en][http://statdb.luke.fi/PXWeb/pxweb/en/LUKE/LUKE__06%20Kala%20ja%20riista__06%20Muut__02%20Kalan%20kulutus/2_Kalankulutus.px/table/tableViewLayout1/?rxid=dc711a9e-de6d-454b-82c2-74ff79a3a5e0] | |||
<t2b name="Fish consumption as food in Finland" index="Origin,Species,Year" locations="1999,2000,2001,2002,2003,2004,2005,2006,2007,2008,2009,2010,2011,2012,2013,2014,2015,2016,2017" unit="kg/a per person"> | |||
domestic fish|Total|6.1|6.1|5.9|6.2|5.8|5.3|5.2|5.0|5.0|4.4|4.5|4.3|3.8|3.8|3.8|4.0|4.1|4.1|4.1 | |||
domestic fish|Farmed rainbow trout|1.6|1.6|1.6|1.6|1.3|1.3|1.4|1.1|1.1|1.2|1.2|1.1|1.0|1.0|1.1|1.1|1.3|1.2|1.2 | |||
domestic fish|Baltic herring|0.8|1.2|1.1|1.1|0.9|0.8|0.7|0.5|0.4|0.4|0.4|0.3|0.3|0.3|0.3|0.3|0.3|0.3|0.3 | |||
domestic fish|Pike|0.8|0.7|0.7|0.7|0.6|0.7|0.7|0.8|0.7|0.6|0.6|0.6|0.5|0.4|0.4|0.5|0.5|0.4|0.4 | |||
domestic fish|Perch|0.7|0.7|0.7|0.7|0.6|0.6|0.6|0.7|0.7|0.5|0.5|0.5|0.4|0.4|0.4|0.5|0.5|0.4|0.4 | |||
domestic fish|Vendace|0.7|0.7|0.7|0.7|0.8|0.8|0.7|0.6|0.6|0.6|0.6|0.7|0.6|0.6|0.6|0.6|0.6|0.5|0.6 | |||
domestic fish|European whitefish|0.4|0.4|0.4|0.4|0.3|0.3|0.3|0.3|0.5|0.3|0.3|0.3|0.3|0.3|0.3|0.3|0.2|0.3|0.3 | |||
domestic fish|Pike perch|0.3|0.2|0.2|0.2|0.3|0.3|0.3|0.3|0.3|0.3|0.3|0.3|0.3|0.3|0.3|0.3|0.3|0.4|0.4 | |||
domestic fish|Other domestic fish|0.8|0.6|0.5|0.8|1.0|0.5|0.5|0.7|0.7|0.5|0.6|0.5|0.4|0.5|0.4|0.5|0.4|0.5|0.5 | |||
imported fish|Total|6.0|6.2|7.0|7.1|8.0|8.6|7.9|8.6|9.7|9.7|9.3|10.2|11.1|10.9|10.8|10.9|10.2|9.1|9.8 | |||
imported fish|Farmed rainbow trout|0.2|0.3|0.4|0.6|0.9|0.6|0.6|0.7|1.0|1.0|0.8|0.8|0.9|1.0|0.9|0.9|0.8|0.9|0.8 | |||
imported fish|Farmed salmon|1.0|0.9|1.2|1.3|1.6|2.2|1.9|2.0|2.7|2.6|2.9|3.1|3.9|4.2|4.0|4.4|4.1|3.5|4.0 | |||
imported fish|Tuna (prepared and preserved)|1.0|1.2|1.4|1.4|1.5|1.6|1.6|1.5|1.7|1.7|1.6|1.7|1.7|1.6|1.9|1.7|1.6|1.4|1.5 | |||
imported fish|Saithe (frozen fillet)|0.7|0.6|0.7|0.7|0.7|0.6|0.4|0.5|0.5|0.5|0.5|0.5|0.6|0.5|0.5|0.5|0.5|0.4|0.4 | |||
imported fish|Shrimps|0.5|0.4|0.5|0.5|0.5|0.5|0.5|0.5|0.6|0.6|0.6|0.7|0.7|0.6|0.6|0.5|0.5|0.4|0.4 | |||
imported fish|Herring and Baltic herring (preserved)|0.6|0.5|0.6|0.6|0.5|0.4|0.5|0.6|0.5|0.6|0.3|0.5|0.4|0.3|0.4|0.5|0.5|0.5|0.5 | |||
imported fish|Other imported fish|2.0|2.3|2.2|2.0|2.3|2.7|2.4|2.8|2.7|2.7|2.6|2.9|2.9|2.7|2.5|2.3|2.3|1.9|2.2 | |||
</t2b> | |||
==== Descriptive statistics ==== | |||
[[File:Goherr survey correlations.png|thumb|300px|Correlation matrix of all questions in the survey (answers converted to numbers).]] | |||
Model must contain predictors such as country, gender, age etc. Maybe we should first study what determinants are important? | |||
Model must also contain determinants that would increase or decrease fish consumption. This should be conditional on the current consumption. How? | |||
Maybe we should look at principal coordinates analysis with all questions to see how they behave. | |||
Also look at correlation table to see clusters. | |||
Some obvious results: | |||
* If reports no fish eating, many subsequent answers are NA. | |||
* No vitamins correlates negatively with vitamin intake. | |||
* Unknown salmon correlates negatively with the types of salmon eaten. | |||
* Different age categories correlate with each other. | |||
However, there are also meaningful negative correlations: | |||
* Country vs allergy | |||
* Country vs Norwegian salmon and Rainbow trout | |||
* Country vs not traditional. | |||
* Country vs recommendation awareness | |||
* Allergy vs economic wellbeing | |||
* Baltic salmon use (4 questions) vs Don't like taste and Not used to | |||
* All questions between Easy to cook ... Traditional dish | |||
* | |||
Meaningful positive correlations: | |||
* All questions between Baltic salmon ... Rainbow trout | |||
* How often Baltic salmon/herring/side salmon/side herring | |||
* How much Baltic salmon/herring/side salmon/side herring | |||
* Better availability ... Recommendation | |||
* All questions between Economic wellbeing...Personal aims | |||
* Omega3, Vitamin D, and Other vitamins | |||
Model runs | |||
* [http://en.opasnet.org/en-opwiki/index.php?title=Special:RTools&id=QaMJZqUX0cPaTfOF Model run 13.3.2017] | |||
* Model run 21.4.2017 [http://en.opasnet.org/en-opwiki/index.php?title=Special:RTools&id=0Lmu6OVAjomgJwAN] old code from Answer merged to this code and debugged | |||
<rcode label="Descriptive statistics" graphics=1> | |||
# This is code Op_en7749/ on page [[Goherr: Fish consumption study]] | |||
library(OpasnetUtils) | |||
library(ggplot2) | |||
library(reshape2) | |||
library(car) | |||
library(vegan) | |||
objects.latest("Op_en7749", "preprocess2") # [[Goherr: Fish consumption study]]: survey, surv | |||
############################# | ############################### From a previous code on Answer | ||
for(i in c(5:6, 16, 29:30, 46:49, 85:86, 95:98, 135)) { | |||
temp <- survey[!is.na(survey[[i]]),] | |||
p <- ggplot(temp, aes(x = temp[[i]])) + geom_bar() + | |||
theme_gray(base_size = 18) + theme(axis.text.x=element_text(angle=90,hjust=1,vjust=0.4)) + | |||
) | labs(title = colnames(temp[i])) + xlab("") + facet_wrap(~ Country) | ||
print(p) | |||
} | |||
temp <- melt(survey, measure.vars = 145:153, variable.name = "Changing.factor", value.name = "Impact") | |||
levs <- c("-5 strongly disagree", "-4", "-3", "-2", "-1", "0 Neutral", "1", "2", "3", "4", "5 strongly agree", "I don't know") | |||
temp$Impact <- factor(temp$Impact, levels = levs, labels = levs, ordered = TRUE) | |||
ggplot(temp, aes(x = Impact, fill = Country)) + geom_bar(position = "dodge") + facet_wrap(~ Changing.factor)+ | |||
theme_gray(base_size = 24) + theme(axis.text.x=element_text(angle=90,hjust=1,vjust=0.4)) | |||
surveytemp <- subset(survey, survey[[31]] == "Yes") | |||
) | |||
temp <- melt(surveytemp, measure.vars = 38:43, variable.name = "Variable", value.name = "Value") | |||
ggplot(temp, aes(x = Value, fill = Country)) + geom_bar(position = "dodge") + facet_wrap(~ Variable)+ | |||
theme_gray(base_size = 24) + labs(title = "Baltic salmon sources") | |||
) | |||
temp <- melt(surveytemp, measure.vars = 50:59, variable.name = "Variable", value.name = "Value") | |||
ggplot(temp, aes(x = Value, fill = Country)) + geom_bar(position = "dodge") + facet_wrap(~ Variable)+ | |||
theme_gray(base_size = 24) + labs(title = "Reasons to eat Baltic salmon") | |||
) | temp <- melt(surveytemp, measure.vars = 73:82, variable.name = "Variable", value.name = "Value") | ||
ggplot(temp, aes(x = Value, fill = Country)) + geom_bar(position = "dodge") + facet_wrap(~ Variable)+ | |||
theme_gray(base_size = 24) + theme(axis.text.x=element_text(angle=90,hjust=1,vjust=0.4)) + labs(title = "Baltic salmon actions") | |||
surveytemp <- subset(survey, survey[[31]] == "No") | |||
) | |||
temp <- melt(surveytemp, measure.vars = 62:70, variable.name = "Variable", value.name = "Value") | |||
ggplot(temp, aes(x = Value, fill = Country)) + geom_bar(position = "dodge") + facet_wrap(~ Variable)+ | |||
theme_gray(base_size = 24) + labs(title = "Reasons not to eat Baltic salmon") | |||
) | |||
surveytemp <- subset(survey, survey[[86]] == "Yes") | |||
) | |||
temp <- melt(surveytemp, measure.vars = 87:92, variable.name = "Variable", value.name = "Value") | |||
ggplot(temp, aes(x = Value, fill = Country)) + geom_bar(position = "dodge") + facet_wrap(~ Variable)+ | |||
theme_gray(base_size = 24) + labs(title = "Baltic herring sources") | |||
temp <- | temp <- melt(surveytemp, measure.vars = 99:108, variable.name = "Variable", value.name = "Value") | ||
ggplot(temp, aes(x = Value, fill = Country)) + geom_bar(position = "dodge") + facet_wrap(~ Variable)+ | |||
theme_gray(base_size = 24) + labs(title = "Reasons to eat Baltic herring") | |||
temp <- melt(surveytemp, measure.vars = 123:132, variable.name = "Variable", value.name = "Value") | |||
ggplot(temp, aes(x = Value, fill = Country)) + geom_bar(position = "dodge") + facet_wrap(~ Variable)+ | |||
theme_gray(base_size = 24) + theme(axis.text.x=element_text(angle=90,hjust=1,vjust=0.4)) + labs(title = "Baltic herring actions") | |||
surveytemp <- subset(survey, survey[[86]] == "No") | |||
melted_correlations <- melt(survey_correlations) | temp <- melt(surveytemp, measure.vars = 111:120, variable.name = "Variable", value.name = "Value") | ||
ggplot(temp, aes(x = Value, fill = Country)) + geom_bar(position = "dodge") + facet_wrap(~ Variable)+ | |||
theme_gray(base_size = 24) + labs(title = "Reasons not to eat Baltic herring") | |||
#####################################3 | |||
temp <- sapply(survey, as.numeric) # Can be done for surv to get a smaller matrix | |||
survey_correlations <- (cor(temp, method="spearman", use="pairwise.complete.obs")) | |||
temp <- colnames(survey_correlations) | |||
melted_correlations <- melt(survey_correlations) | |||
melted_correlations$Var1 <- factor(melted_correlations$Var1, levels=temp) | melted_correlations$Var1 <- factor(melted_correlations$Var1, levels=temp) | ||
| Line 1,137: | Line 1,433: | ||
scale_fill_gradient2(low = "#480610", mid = "#FFFFFF", high = "#06480F", midpoint = 0, space = "Lab", guide = "colourbar") | scale_fill_gradient2(low = "#480610", mid = "#FFFFFF", high = "#06480F", midpoint = 0, space = "Lab", guide = "colourbar") | ||
####################### | ####################### Descriptive statistics | ||
oprint(cor(surv, use = "pairwise.complete.obs")) | |||
# --> Baltic salmon and herring eating are correlated, so they should be estimated together | |||
oprint(table(surv[c(12,7,4)], useNA = "ifany")) | |||
oprint(table(surv[c(13,12)], useNA = "ifany")) | |||
## | ############################# Plot original data | ||
# Eating frequencies of fish and Baltic salmon and herring with random noise, all | |||
pl <- surv[surv$Eatfish,c(5,8,10,13,15)] | |||
scatterplotMatrix( | |||
data.matrix(pl) + runif(nrow(pl)*ncol(pl), -0.5, 0.5) | |||
) | |||
ggplot(data.frame( | |||
X = rep(1,5), Y = 5:1, | |||
legend = c("All", "Finland", "Sweden", "Denmark", "Estonia") | |||
), aes(x=X, y=Y, label=legend))+ | |||
geom_text()+ | |||
labs(title="Fish eating questions with random noise") | |||
## Fish eating questions with some random noise, all | |||
pl <- surv[surv$Eatfish,c(5,6,7,12)] | |||
scatterplotMatrix( | |||
data.matrix(pl) + runif(nrow(pl)*ncol(pl), -0.5, 0.5) | |||
) | |||
* | |||
< | ## Fish eating questions with some random noise, FI | ||
pl <- surv[surv$Country == 1 & surv$Eatfish,c(5,6,7,12)] | |||
scatterplotMatrix( | |||
data.matrix(pl) + runif(nrow(pl)*ncol(pl), -0.5, 0.5) | |||
) | |||
## Fish eating questions with some random noise, SWE | |||
pl <- surv[surv$Country == 2 & surv$Eatfish,c(5,6,7,12)] | |||
scatterplotMatrix( | |||
data.matrix(pl) + runif(nrow(pl)*ncol(pl), -0.5, 0.5) | |||
) | |||
# Fish | ## Fish eating questions with some random noise, Dk | ||
pl <- surv[surv$Country == 3 & surv$Eatfish,c(5,6,7,12)] | |||
scatterplotMatrix( | |||
data.matrix(pl) + runif(nrow(pl)*ncol(pl), -0.5, 0.5) | |||
) | |||
## Fish eating questions with some random noise, EST | |||
pl <- surv[surv$Country == 4 & surv$Eatfish,c(5,6,7,12)] | |||
scatterplotMatrix( | |||
data.matrix(pl) + runif(nrow(pl)*ncol(pl), -0.5, 0.5) | |||
) | |||
## Baltic herring questions with some random noise | |||
pl <- surv[surv$Eatherr,13:16] | |||
scatterplotMatrix( | |||
data.matrix(pl) + runif(nrow(pl)*ncol(pl), -0.5, 0.5) | |||
) | |||
## Baltic salmon questions with some random noise | |||
pl <- surv[surv$Eatsalm,8:11] | |||
scatterplotMatrix( | |||
data.matrix(pl) + runif(nrow(pl)*ncol(pl), -0.5, 0.5) | |||
) | |||
##################### CORRELATION MATRIX | |||
temp <- sapply(survey, as.numeric) # Can be done for surv to get a smaller matrix | |||
survey_correlations <- (cor(temp, method="spearman", use="pairwise.complete.obs")) | |||
temp <- colnames(survey_correlations) | |||
melted_correlations <- melt(survey_correlations) | |||
melted_correlations$Var1 <- factor(melted_correlations$Var1, levels=temp) | |||
melted_correlations$Var2 <- factor(melted_correlations$Var2, levels=temp) | |||
melted_correlations$value <- ifelse(melted_correlations$value >= 0.99,NA,melted_correlations$value) | |||
ggplot(melted_correlations, aes(x = Var1, y = Var2, fill = value, label= round(value, 2)))+ | |||
geom_raster()+ | |||
theme(axis.text.x=element_text(angle=90,hjust=1,vjust=0.4))+ | |||
scale_fill_gradient2(low = "#480610", mid = "#FFFFFF", high = "#06480F", midpoint = 0, space = "Lab", guide = "colourbar") | |||
############################### PRINCIPAL COORDINATE ANALYSIS (PCoA) | |||
#tämä osa valmistaa sen datan. | |||
hypocols1 <- c(46:49,95:98) | |||
answ <- sapply(survey[hypocols1], FUN=as.numeric) | |||
answ <- as.matrix(answ[!is.na(rowSums(answ)),]) | |||
pcoa_caps <- capscale(t(answ) ~ 1, distance="euclidean") ##PCoA done | |||
) | |||
## Kuva koko hypoteeseista | |||
colstr <- c("palevioletred1","royalblue1","seagreen1","violet","khaki2","skyblue", "orange") | |||
#hypo_sizes <- (5 - colMeans(answ)) | |||
# | #leg_sizes <- c(4, 3, 2, 1, 0.01) | ||
#pdf(file="pcoa_plot.pdf", height=6, width=7.5) | |||
plot(pcoa_caps, display = c("sp", "wa"), type="n")#, xlim=c(-6,4.5)) ## PCoA biplot, full scale | |||
points(pcoa_caps, display= c("sp"), col="gray40") # adding the people points | |||
points(pcoa_caps, display= c("wa"), pch=19)#, cex=hypo_sizes, col=trait.cols) | |||
) | text(pcoa_caps, display=c("wa"), srt=25, cex=0.5) | ||
#legend(x=-6, y=3.8, levels(traits), fill=colstr, bty="n", cex=1) | |||
#legend(-6, -2, legend=c("Very likely", "Moderately likely", | |||
# "No opinion", "Moderately unlikely", "Very unlikely"), | |||
# pch=21, pt.cex = leg_sizes, bty="n", cex=1) | |||
#dev.off() | |||
</rcode> | |||
==== Bayes model ==== | |||
* Model run 3.3.2017. All variables assumed independent. [http://en.opasnet.org/en-opwiki/index.php?title=Special:RTools&id=lwlSwXazIDHDyJJg] | |||
* Model run 3.3.2017. p has more dimensions. [http://en.opasnet.org/en-opwiki/index.php?title=Special:RTools&id=ZmbNUuZeb7UOf8NP] | |||
* Model run 25.3.2017. Several model versions: strange binomial+multivarnormal, binomial, fractalised multivarnormal [http://en.opasnet.org/en-opwiki/index.php?title=Special:RTools&id=pKe0s2Ldm1mbIVuO] | |||
* Model run 27.3.2017 [http://en.opasnet.org/en-opwiki/index.php?title=Special:RTools&id=2hY9p2r8CTJi3Qwq] | |||
* Other models except multivariate normal were [http://en.opasnet.org/en-opwiki/index.php?title=Goherr:_Fish_consumption_study&oldid=40185 archived] and removed from active code 29.3.2017. | |||
* Model run 29.3.2017 with raw data graphs [http://en.opasnet.org/en-opwiki/index.php?title=Special:RTools&id=BB8nePJb7hzSw6Ha] | |||
* Model run 29.3.2017 with salmon and herring ovariables stored [http://en.opasnet.org/en-opwiki/index.php?title=Special:RTools&id=2Hz4tYjrQLnUfIXw] | |||
* Model run 13.4.2017 with first version of coordinate matrix and principal coordinate analysis [http://en.opasnet.org/en-opwiki/index.php?title=Special:RTools&id=2k2dKhYPc2UkOCY5] | |||
* Model run 20.4.2017 [http://en.opasnet.org/en-opwiki/index.php?title=Special:RTools&id=QXRSZOliFNfJil39] code works but needs a safety check against outliers | |||
* Model run 21.4.2017 [http://en.opasnet.org/en-opwiki/index.php?title=Special:RTools&id=xbt2OoXPG8fi5COL] some model results plotted | |||
* Model run 21.4.2017 [http://en.opasnet.org/en-opwiki/index.php?title=Special:RTools&id=waDgScuIkUvEShLy] ovariables produced by the model stored. | |||
* Model run 18.5.2017 [http://en.opasnet.org/en-opwiki/index.php?title=Special:RTools&id=hy2clSt3Vo5y4xN5] small updates | |||
* 13.2.2018 old model run but with new Opasnet [http://en.opasnet.org/en-opwiki/index.php?title=Special:RTools&id=flr56viVhZ0VzZQu] | |||
<rcode name="bayes" label="Initiate Bayes model (for developers only)" graphics=1> | |||
# This is code Op_en7749/bayes on page [[Goherr: Fish consumption study]] | |||
library(OpasnetUtils) | |||
library(ggplot2) | |||
library(reshape2) | |||
library(rjags) | |||
library(car) | |||
library(vegan) | |||
library(MASS) | |||
#library(gridExtra) # Error: package ‘gridExtra’ was built before R 3.0.0: please re-install it | |||
# Fish intake in humans | |||
# Data from data.frame survey from page [[Goherr: Fish consumption study]] | |||
objects.latest("Op_en7749", "preprocess2") # [[Goherr: Fish consumption study]]: survey, surv, ... | |||
cat("Version with multivariate normal.\n") | |||
# Development needs: | |||
# | ## Correlation between salmon.often and herring.often needs to be estimated. | ||
## Gender, country and age-spesific values should be estimated. | |||
mod <- textConnection(" | |||
model{ | |||
for(i in 1:S) { | |||
survs[i,1:4] ~ dmnorm(mus[], Omegas[,]) | |||
} | |||
for(j in 1:H) { | |||
survh[j,1:4] ~ dmnorm(muh[], Omegah[,]) | |||
} | |||
mus[1:4] ~ dmnorm(mu0[1:4], S2[1:4,1:4]) | |||
Omegas[1:4,1:4] ~ dwish(S3[1:4,1:4],S) | |||
anss.pred ~ dmnorm(mus[], Omegas[,]) | |||
muh[1:4] ~ dmnorm(mu0[1:4], S2[1:4,1:4]) | |||
Omegah[1:4,1:4] ~ dwish(S3[1:4,1:4],H) | |||
) | ansh.pred ~ dmnorm(muh[], Omegah[,]) | ||
} | |||
") | |||
jags <- jags.model( | |||
mod, | |||
data = list( | |||
survs = surv[surv$Eatsalm,c(8:11)], | |||
S = sum(surv$Eatsalm), | |||
survh = surv[surv$Eatherr,c(13:16)], | |||
H = sum(surv$Eatherr), | |||
mu0 = rep(2,4), | |||
S2 = diag(4)/100000, | |||
S3 = diag(4)/10000 | |||
), | |||
n.chains = 4, | |||
n.adapt = 100 | |||
) | ) | ||
update(jags, 100) | |||
samps.j <- jags.samples( | |||
< | jags, | ||
c('mus', 'Omegas', 'anss.pred','muh','Omegah','ansh.pred'), | |||
1000 | |||
) | |||
js <- array( | |||
c( | |||
samps.j$mus[,,1], | |||
samps.j$Omegas[,1,,1], | |||
samps.j$Omegas[,2,,1], | |||
samps.j$Omegas[,3,,1], | |||
samps.j$Omegas[,4,,1], | |||
samps.j$anss.pred[,,1], | |||
samps.j$muh[,,1], | |||
samps.j$Omegah[,1,,1], | |||
samps.j$Omegah[,2,,1], | |||
samps.j$Omegah[,3,,1], | |||
samps.j$Omegah[,4,,1], | |||
samps.j$ansh.pred[,,1] | |||
), | |||
dim = c(4,1000,6,2), | |||
dimnames = list( | |||
Question = 1:4, | |||
Iter = 1:1000, | |||
Parameter = c("mu","Omega1", "Omega2", "Omega3", "Omega4", "ans.pred"), | |||
Fish = c("Salmon", "Herring") | |||
) | |||
) | |||
# Mu for all questions about salmon | |||
scatterplotMatrix(t(js[,,1,1])) | |||
# All parameters for question 1 about salmon | |||
scatterplotMatrix(js[1,,,1]) | |||
# Same for herring | |||
scatterplotMatrix(js[1,,,2]) | |||
jsd <- melt(js) | |||
#ggplot(jsd, aes(x=value, colour=Question))+geom_density()+facet_grid(Parameter ~ Fish) | |||
#ggplot(as.data.frame(js), aes(x = anss.pred, y = Sampled))+geom_point()+stat_ellipse() | |||
coda.j <- coda.samples( | |||
jags, | |||
c('mus', 'Omegas', 'anss.pred'), | |||
1000 | |||
) | |||
plot(coda.j) | |||
######## fish.param contains expected values of the distribution parameters from the model | |||
fish.param <- list( | |||
mu = apply(js[,,1,], MARGIN = c(1,3), FUN = mean), | |||
Omega = lapply( | |||
1:2, | |||
FUN = function(x) { | |||
solve(apply(js[,,2:5,], MARGIN = c(1,3,4), FUN = mean)[,,x]) | |||
} # solve matrix: precision->covariace | |||
) | |||
) | |||
objects.store(fish.param) | |||
cat("List fish.param stored.\n") | |||
</rcode> | |||
==== Initiate ovariables ==== | |||
sur | |||
=====jsp taken directly from data WITHOUT salmpling===== | |||
<rcode name="nonsamplejsp" label="Initiate jsp from data without sampling (for developers only)" embed=1> | |||
# This is code Op_en7749/nonsamplejsp on page [[Goherr: Fish consumption study]] | |||
# The code produces amount esimates (jsp ovariable) directly from data rather than bayesian model or sampling. | |||
library(OpasnetUtils) | |||
jsp <- Ovariable( | |||
"jsp", | |||
dependencies = data.frame(Name="survey1", Ident="Op_en7749/preprocess2"), # [[Goherr: Fish consumption study]] | |||
formula = function(...) { | |||
require(reshape2) | |||
sur <- survey1[c(157,1,3,158,162:178,81,82,131,132,154)] # Removed, not needed:16,29,30? | |||
# | #colnames(sur) | ||
#[1] "Row" "Country" "Gender" | |||
#[4] "Ages" "Baltic.salmon.n" "How.often.BS.n" | |||
"Less.chemicals" | #[7] "How.much.BS.n" "How.often.side.BS.n" "How.much.side.BS.n" | ||
"Recommended" | #[10] "Better.availability.BS.n" "Less.chemicals.BS.n" "Eat.BH.n" | ||
#[13] "How.often.BH.n" "How.much.BH.n" "How.often.side.BH.n" | |||
#[16] "How.much.side.BH.n" "Better.availability.BH.n" "Less.chemicals.BH.n" | |||
sur | #[19] "Eatfish" "Eatsalm" "Eatherr" | ||
#[22] "Recommended.BS" "Not.recommended.BS" "Recommended.BH" | |||
#[25] "Not.recommended.BH" "Weighting" | |||
# Make sure that Row is kept separate from Iter because in the sampling version they are different. | |||
# sur contained columns Eat.fish, How.often.fish, Eat.salmon. Are these needed, as all other questions are melted? No | |||
# Columns 5-7 removed, so colnames list above does not match. | |||
return(Ovariable( | colnames(sur)[5] <- "Eat.BS" | ||
output = sur, | colnames(sur) <- gsub("\\.n","",colnames(sur)) | ||
marginal = colnames(sur) %in% c("Fish", "Iter", "Question") | sur[22:25] <- sapply(sur[22:25], as.numeric) | ||
)) | sur <- melt( | ||
} | sur, | ||
id.vars=c(1:4,26), | |||
variable.name="Question", | |||
value.name="Result" | |||
) | |||
sur$Fish <- ifelse(grepl("BH",sur$Question),"Herring","Salmon") | |||
sur$Question <- gsub("\\.BS","",sur$Question) | |||
sur$Question <- gsub("\\.BH","",sur$Question) | |||
########## If the missing values are not adjusted, they drop out in the next stage. | |||
if(TRUE) { | |||
# The adjustments below probably should go to the preprocess2 code. | |||
sur$Result[sur$Question %in% c( | |||
"Better.availability", | |||
"Less.chemicals", | |||
"Recommended", | |||
"Not.recommended" | |||
) & is.na(sur$Result)] <- 4 # If missing --> no change | |||
sur$Result[is.na(sur$Result)] <- 1 # Replace missing values with 1. That will produce 0 g/d. | |||
} | |||
return(Ovariable( | |||
output = sur, | |||
marginal = colnames(sur) %in% c("Fish", "Iter", "Question") | |||
)) | |||
} | |||
) | ) | ||
objects.store(jsp) | objects.store(jsp) | ||
cat("Ovariable jsp with actual survey data: | cat("Ovariable jsp with actual survey data: every respondent is kept in data without sampling.\n") | ||
</rcode> | </rcode> | ||
===== Amount estimated from a bayesian model ===== | |||
* Model run 24.5.2017 [http://en.opasnet.org/en-opwiki/index.php?title=Special:RTools&id=3UORzPwospQxp82h] | |||
<rcode name="modeljsp" label="Initiate jsp from model (for developers only)"> | |||
# This is code Op_en7749/modeljsp on page [[Goherr: Fish consumption study]] | |||
<rcode name=" | |||
# This is code Op_en7749/ | |||
library(OpasnetUtils) | library(OpasnetUtils) | ||
jsp <- Ovariable( | |||
"jsp", | |||
dependencies = data.frame(Name = "fish.param", Ident = "Op_en7749/bayes"), | |||
" | |||
dependencies = data.frame( | |||
formula = function(...) { | formula = function(...) { | ||
require(MASS) | |||
require(reshape2) | |||
jsp <- lapply( | |||
1:2, | |||
FUN = function(x) { | |||
mvrnorm(openv$N, fish.param$mu[,x], fish.param$Omega[[x]]) | |||
} | |||
) | |||
jsp <- rbind( | |||
cbind( | |||
Fish = "Salmon", | |||
Iter = 1:nrow(jsp[[1]]), | |||
as.data.frame(jsp[[1]]) | |||
), | |||
cbind( | |||
Fish = "Herring", | |||
Iter = 1:nrow(jsp[[2]]), | |||
as.data.frame(jsp[[2]]) | |||
) | |||
) | ) | ||
jsp <- melt(jsp, id.vars = c("Iter", "Fish"), variable.name = "Question", value.name = "Result") | |||
colnames( | jsp <- Ovariable(output=jsp, marginal = colnames(jsp) %in% c("Iter", "Fish", "Question")) | ||
return( | return(jsp) | ||
} | } | ||
) | ) | ||
objects.store(jsp) | |||
cat("Ovariable jsp stored.\n") | |||
</rcode> | |||
===== Amount estimates directly from data rather than from a bayesian model ===== | |||
* Initiation run 18.5.2017 [http://en.opasnet.org/en-opwiki/index.php?title=Special:RTools&id=crW1kboP72BN1JbK] | |||
* Initiation run 24.2.2018: sampling from survey rather than each respondent once [http://en.opasnet.org/en-opwiki/index.php?title=Special:RTools&id=yJavDqd5zPUyt5pK] | |||
<rcode name="surveyjsp" label="Initiate jsp from survey (for developers only)" embed=1> | |||
" | # This is code Op_en7749/surveyjsp on page [[Goherr: Fish consumption study]] | ||
dependencies = data.frame( | # The code produces amount esimates (jsp ovariable) directly from data rather than bayesian model. | ||
library(OpasnetUtils) | |||
jsp <- Ovariable( | |||
"jsp", | |||
dependencies = data.frame(Name="survey1", Ident="Op_en7749/preprocess2"), # [[Goherr: Food cunsumption study]] | |||
formula = function(...) { | formula = function(...) { | ||
require(reshape2) | |||
sur <- survey1[ | |||
sample(1:nrow(survey1), openv$N, replace=TRUE, prob=survey1$Weighting) , | |||
c(157,1,3,158,162:178,81,82,131,132) # Removed, not needed:16,29,30? | |||
] | |||
sur$Iter <- 1:openv$N | |||
#colnames(sur) | |||
#[1] "Row" "Country" "Gender" | |||
#[4] "Ages" "Baltic.salmon.n" "How.often.BS.n" | |||
#[7] "How.much.BS.n" "How.often.side.BS.n" "How.much.side.BS.n" | |||
#[10] "Better.availability.BS.n" "Less.chemicals.BS.n" "Eat.BH.n" | |||
#[13] "How.often.BH.n" "How.much.BH.n" "How.often.side.BH.n" | |||
#[16] "How.much.side.BH.n" "Better.availability.BH.n" "Less.chemicals.BH.n" | |||
#[19] "Eatfish" "Eatsalm" "Eatherr" | |||
#[22] "Recommended.BS" "Not.recommended.BS" "Recommended.BH" | |||
#[25] "Not.recommended.BH" "Iter" | |||
# Make sure that Row is kept separate from Iter because in the sampling version they are different. | |||
# sur contained columns Eat.fish, How.often.fish, Eat.salmon. Are these needed, as all other questions are melted? No | |||
# Columns 5-7 removed, so colnames list above does not match. | |||
colnames(sur)[5] <- "Eat.BS" | |||
colnames(sur) <- gsub("\\.n","",colnames(sur)) | |||
sur[22:25] <- sapply(sur[22:25], as.numeric) | |||
sur <- melt( | |||
sur, | |||
id.vars=c(1:4,26), | |||
variable.name="Question", | |||
value.name="Result" | |||
) | ) | ||
sur$Fish <- ifelse(grepl("BH",sur$Question),"Herring","Salmon") | |||
sur$Question <- gsub("\\.BS","",sur$Question) | |||
sur$Question <- gsub("\\.BH","",sur$Question) | |||
} | |||
) | # The adjustments below probably should go to the preprocess2 code. | ||
sur$Result[sur$Question %in% c( | |||
"Better.availability", | |||
"Less.chemicals", | |||
"Recommended", | |||
"Not.recommended" | |||
) & is.na(sur$Result)] <- 4 # If missing --> no change | |||
sur$Result[is.na(sur$Result)] <- 1 # Replace missing values with 1. That will produce 0 g/d. | |||
return(Ovariable( | |||
output = sur, | |||
marginal = colnames(sur) %in% c("Fish", "Iter", "Question") | |||
)) | |||
} | |||
) | |||
objects.store(jsp) | |||
cat("Ovariable jsp with actual survey data: a respondent is sampled for each iteration.\n") | |||
</rcode> | |||
===== Initiate other ovariables ===== | |||
* Code stores ovariables assump, often, much, oftenside, muchside, amount. | |||
" | * Model run 19.5.2017 [http://en.opasnet.org/en-opwiki/index.php?title=Special:RTools&id=FauP3x6hviC4xWCD] | ||
dependencies = data.frame( | * Initiation run 24.5.2017 without jsp [http://en.opasnet.org/en-opwiki/index.php?title=Special:RTools&id=6U3FWqVRaNumPcok] | ||
Name=c("jsp","assump"), | * Model run 8.6.2017 [http://en.opasnet.org/en-opwiki/index.php?title=Special:RTools&id=euGolhOQvPqzQDvf] | ||
<rcode name="initiate" label="Initiate ovariables (for developers only)" embed=1> | |||
# This is code Op_en7749/initiate on page [[Goherr: Fish consumption study]] | |||
library(OpasnetUtils) | |||
# Combine modelled survey answers with estimated amounts and frequencies by: | |||
# Rounding the modelled result and merging that with value in ovariable assump | |||
often <- Ovariable( | |||
"often", | |||
dependencies = data.frame( | |||
Name=c("jsp","assump"), | |||
Ident=c("Op_en7749/surveyjsp","Op_en7749/initiate") | Ident=c("Op_en7749/surveyjsp","Op_en7749/initiate") | ||
), | ), | ||
formula = function(...) { | formula = function(...) { | ||
out <- jsp[jsp$Question == "How.often" , ] | |||
out$Value <- round(result(out)) | |||
out <- merge( | |||
assump@output[assump$Variable == "freq",], | |||
out@output | |||
assump@output[assump$Variable == " | |||
) | ) | ||
out <- out[!colnames(out) %in% c("Value", "Variable", "Result","Question")] | |||
colnames(out)[colnames(out) == "assumpResult"] <- "Result" | |||
return(out) | |||
colnames( | |||
return( | |||
} | } | ||
) | ) | ||
much <- Ovariable( | |||
" | "much", | ||
dependencies = data.frame( | dependencies = data.frame( | ||
Name=c("jsp","assump"), | Name=c("jsp","assump"), | ||
| Line 1,554: | Line 1,907: | ||
), | ), | ||
formula = function(...) { | formula = function(...) { | ||
out <- jsp[jsp$Question == "How.much" , ] | |||
out$Value <- round(result(out)) | |||
out <- merge( | |||
assump@output[assump$Variable == "amdish",], | |||
assump@output[assump$Variable == " | out@output | ||
) | ) | ||
colnames(o1)[colnames(o1)=="assumpResult"] <- "Result" | out <- out[!colnames(out) %in% c("Value", "Variable", "Result","Question")] | ||
o1$Recommendation <- "Eat more" | colnames(out)[colnames(out) == "assumpResult"] <- "Result" | ||
return(out) | |||
# Recommend less fish | } | ||
o2 <- jsp[jsp$Question == "Not.recommended" , ] | ) | ||
o2$Value <- round(result(o2)) | |||
o2 <- merge( | oftenside <- Ovariable( | ||
assump@output[assump$Variable == "change",], | "oftenside", | ||
o2@output | dependencies = data.frame( | ||
Name=c("jsp","assump"), | |||
Ident=c("Op_en7749/surveyjsp","Op_en7749/initiate") | |||
), | |||
formula = function(...) { | |||
out <- jsp[jsp$Question == "How.often.side" , ] | |||
out$Value <- round(result(out)) | |||
out <- merge( | |||
assump@output[assump$Variable == "freq",], | |||
out@output | |||
) | |||
out <- out[!colnames(out) %in% c("Value", "Variable", "Result","Question")] | |||
colnames(out)[colnames(out) == "assumpResult"] <- "Result" | |||
return(out) | |||
} | |||
) | |||
muchside <- Ovariable( | |||
"muchside", | |||
dependencies = data.frame( | |||
Name=c("jsp","assump"), | |||
Ident=c("Op_en7749/surveyjsp","Op_en7749/initiate") | |||
), | |||
formula = function(...) { | |||
out <- jsp[jsp$Question == "How.much.side" , ] | |||
out$Value <- round(result(out)) | |||
out <- merge( | |||
assump@output[assump$Variable == "amside",], | |||
out@output | |||
) | |||
out <- out[!colnames(out) %in% c("Value", "Variable", "Result","Question")] | |||
colnames(out)[colnames(out) == "assumpResult"] <- "Result" | |||
return(out) | |||
} | |||
) | |||
assump <- Ovariable( | |||
"assump", | |||
ddata = "Op_en7749", subset = "Assumptions for calculations" | |||
) | |||
effinfo <- Ovariable( # Effect of information | |||
"effinfo", | |||
dependencies = data.frame( | |||
Name=c("jsp","assump"), | |||
Ident=c("Op_en7749/surveyjsp","Op_en7749/initiate") | |||
), | |||
formula = function(...) { | |||
away <- c("Value","Variable","Result","Question","assumpUnit","assumpSource","jspResult","jspSource") | |||
# Better availability of fish | |||
o1 <- jsp[jsp$Question == "Better.availability" , ] | |||
o1$Value <- round(result(o1)) | |||
o1 <- merge( | |||
assump@output[assump$Variable == "change",], | |||
o1@output | |||
) | |||
colnames(o1)[colnames(o1)=="assumpResult"] <- "Better.availabilityResult" | |||
o1 <- Ovariable(name="Better.availability", output=o1[!colnames(o1) %in% away]) | |||
# Less chemicals | |||
o2 <- jsp[jsp$Question == "Less.chemicals" , ] | |||
o2$Value <- round(result(o2)) | |||
o2 <- merge( | |||
assump@output[assump$Variable == "change",], | |||
o2@output | |||
) | |||
colnames(o2)[colnames(o2)=="assumpResult"] <- "Less.chemicalsResult" | |||
o2 <- Ovariable(name="Less.chemicals", output=o2[!colnames(o2) %in% away]) | |||
out <- (o1 + o2)@output | |||
return(Ovariable(output=out, marginal=colnames(out) %in% c("Iter","Fish"))) | |||
} | |||
) | |||
effrecommRaw <- Ovariable( # Effect of recommendations | |||
"effrecommRaw", | |||
dependencies = data.frame( | |||
Name=c("jsp","assump"), | |||
Ident=c("Op_en7749/surveyjsp","Op_en7749/initiate") | |||
), | |||
formula = function(...) { | |||
# Recommend more fish | |||
o1 <- jsp[jsp$Question == "Recommended" , ] | |||
o1$Value <- round(result(o1)) | |||
o1 <- merge( | |||
assump@output[assump$Variable == "change",], | |||
o1@output | |||
) | |||
colnames(o1)[colnames(o1)=="assumpResult"] <- "Result" | |||
o1$Recommendation <- "Eat more" | |||
# Recommend less fish | |||
o2 <- jsp[jsp$Question == "Not.recommended" , ] | |||
o2$Value <- round(result(o2)) | |||
o2 <- merge( | |||
assump@output[assump$Variable == "change",], | |||
o2@output | |||
) | ) | ||
colnames(o2)[colnames(o2)=="assumpResult"] <- "Result" | colnames(o2)[colnames(o2)=="assumpResult"] <- "Result" | ||
o2$Recommendation <- "Eat less" | o2$Recommendation <- "Eat less" | ||
out <- rbind(o1, o2)[!colnames(o1) %in% | out <- rbind(o1, o2)[!colnames(o1) %in% | ||
c("Value","Variable","Question","assumpUnit","assumpSource","jspResult","jspSource") | c("Value","Variable","Question","assumpUnit","assumpSource","jspResult","jspSource") | ||
] | ] | ||
return(Ovariable(output=out, marginal=colnames(out) %in% c("Iter","Fish","Recommendation"))) | |||
}, | |||
unit = "change of amount in fractions" | |||
) | |||
# effrecommRaw and effrecomm are needed to enable decisions to affect the effect of recommendation before | |||
# the ovariable is collapsed. So, target the decisions to effrecommRaw and collapses to effrecomm. | |||
effrecomm <- Ovariable( | |||
"effrecomm", | |||
dependencies = data.frame(Name="effrecommRaw"), | |||
formula = function(...) { | |||
return(effrecommRaw) | |||
} | |||
) | |||
amountRaw <- Ovariable( | |||
"amountRaw", | |||
dependencies = data.frame(Name = c( | |||
"often", | |||
"much", | |||
"oftenside", | |||
"muchside", | |||
"assump" | |||
)), | |||
formula = function(...) { | |||
away <- c( | |||
"assumpUnit", | |||
"jspResult", | |||
"jspSource", | |||
"Variable", | |||
"Value", | |||
"Explanation" | |||
) | |||
often <- often[ , !colnames(often@output) %in% away] | |||
much <- much[ , !colnames(much@output) %in% away] | |||
oftenside <- oftenside[ , !colnames(oftenside@output) %in% away] | |||
muchside <- muchside[ , !colnames(muchside@output) %in% away] | |||
assump <- assump[assump$Variable == "ingredient", !colnames(assump@output) %in% away] | |||
out <- (often * much + oftenside * muchside * assump)/365 # g /d | |||
return(out) | |||
} | |||
) | |||
amount <- Ovariable( | |||
"amount", | |||
dependencies = data.frame(Name = c( | |||
"amountRaw", | |||
"effinfo", | |||
"effrecomm" | |||
)), | |||
formula = function(...) { | |||
out <- amountRaw * (1 + effinfo + effrecomm) | |||
result(out)[result(out)<0] <- 0 | |||
result(out)[result(out)>100] <- 100 | |||
return(out) | |||
} | |||
) | |||
rm(wiki_username) | |||
objects.store(list=ls()) | |||
cat("Ovariables", ls(), "stored.\n") | |||
</rcode> | |||
==== Other code ==== | |||
This is code for analysing EFSA food intake data about fish for BONUS GOHERR manuscript Ronkainen L, Lehikoinen A, Haapasaari P, Tuomisto JT. 2019. | |||
<rcode> | |||
################################ | |||
# This is code for analysing EFSA food intake data about fish for manuscript | |||
# Ronkainen L, Lehikoinen A, Haapasaari P, Tuomisto JT. 2019 | |||
library(ggplot2) | |||
# Change the encoding from UCS-2 to UTF-8 first. | |||
dat <- read.csv( | |||
# "C:/_Eivarmisteta/EFSA fish chronic intake.csv", | |||
"C:/_Eivarmisteta/EFSA herring salmon chronic intake.csv", | |||
header=TRUE, sep=",",dec=".",quote='\"' | |||
) | ) | ||
#> colnames(dat) | |||
#[1] "ï..Survey.s.country" "Survey.start.year" "Survey" | |||
#[4] "Population.Group..L2." "pop.order" "Exposure.hierarchy..L1." | |||
" | #[7] "Number.of.subjects" "Number.of.consumers" "Mean" | ||
#[10] "Standard.Deviation" "X5th.percentile" "X10th.percentile" | |||
#[13] "Median" "X95th.percentile" "X97.5th.percentile" | |||
#[16] "X99th.percentile" "Comment" | |||
colnames(dat)[c(1,2,4,12)] <- c("Country","Year","Group","Fish") | |||
#> levels(dat$Group) | |||
#[1] "Adolescents" "Adults" "Elderly" "Infants" "Lactating women" | |||
#[6] "Other children" "Pregnant women" "Toddlers" "Very elderly" | |||
levels(dat$Group) <- c("Children","Adults","Elderly","Children","Pregnant women", | |||
"Children","Pregnant women","Children","Elderly") | |||
dat <- dat[dat$Fish !="Diadromous fish",] # Foodex2 level 7 entries | |||
ggplot(dat,aes(x=Country, y=Mean, colour=Group))+ | |||
geom_point()+coord_flip()+facet_wrap(~Fish, scales="free")+ | |||
labs(title="Fish consumption in European countries", | |||
y="Average consumption (g/d)") | |||
</rcode> | </rcode> | ||
Latest revision as of 15:39, 12 August 2020
In Opasnet many pages being worked on and are in different classes of progression. Thus the information on those pages should be regarded with consideration. The progression class of this page has been assessed:
|
The content and quality of this page is/was being curated by the project that produced the page.
The quality was last checked: 2019-08-26. |
This page is a study.
The page identifier is Op_en7749 |
|---|
| Moderator:Arja (see all) |
|
|
| Upload data
|
- This page contains a detailed description about data management and analysis of an international survey related to scientific article Forage Fish as Food: Consumer Perceptions on Baltic Herring by Mia Pihlajamäki, Arja Asikainen, Suvi Ignatius, Päivi Haapasaari, and Jouni T. Tuomisto.[1] The results of this survey where also used in another article Health effects of nutrients and environmental pollutants in Baltic herring and salmon: a quantitative benefit-risk assessment by the same group.[2]
Question
How Baltic herring and salmon are used as human food in Baltic sea countries? Which determinants affect on people’s eating habits of these fish species?
Answer
- Model run with all the results of the article[1] 26.8.2018
- Original questionnaire analysis results 13.3.2017
- Consumption amount estimates
- Model run 21.4.2017 [1] first distribution
- Model run 18.5.2017 with modelled data; with direct survey data
Rationale
Survey of eating habits of Baltic herring and salmon in Denmark, Estonia, Finland and Sweden has been done in September 2016 by Taloustutkimus oy. Content of the questionnaire can be accessed in Google drive. The actual data can be found from the link below (see Data).
Data
Questionnaire
Original datafile File:Goherr fish consumption.csv.
| Show details | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Assumptions
The following assumptions are used:
| Obs | Variable | Value | Unit | Result | Description |
|---|---|---|---|---|---|
| 1 | freq | 1 | times /a | 0 | Never |
| 2 | freq | 2 | times /a | 0.5 - 0.9 | less than once a year |
| 3 | freq | 3 | times /a | 2 - 5 | A few times a year |
| 4 | freq | 4 | times /a | 12 - 36 | 1 - 3 times per month |
| 5 | freq | 5 | times /a | 52 | once a week |
| 6 | freq | 6 | times /a | 104 - 208 | 2 - 4 times per week |
| 7 | freq | 7 | times /a | 260 - 364 | 5 or more times per week |
| 8 | amdish | 1 | g /serving | 20 - 70 | 1/6 plate or below (50 grams) |
| 9 | amdish | 2 | g /serving | 70 - 130 | 1/3 plate (100 grams) |
| 10 | amdish | 3 | g /serving | 120 - 180 | 1/2 plate (150 grams) |
| 11 | amdish | 4 | g /serving | 170 - 230 | 2/3 plate (200 grams) |
| 12 | amdish | 5 | g /serving | 220 - 280 | 5/6 plate (250 grams) |
| 13 | amdish | 6 | g /serving | 270 - 400 | full plate (300 grams) |
| 14 | amdish | 7 | g /serving | 400 - 550 | overly full plate (500 grams) |
| 15 | ingredient | fraction | 0.1 - 0.3 | Fraction of fish in the dish | |
| 16 | amside | 1 | g /serving | 20 - 70 | 1/6 plate or below (50 grams) |
| 17 | amside | 2 | g /serving | 70 - 130 | 1/4 plate (100 grams) |
| 18 | amside | 3 | g /serving | 120 - 180 | 1/2 plate (150 grams) |
| 19 | amside | 4 | g /serving | 170 - 230 | 2/3 plate (200 grams) |
| 20 | amside | 5 | g /serving | 220 - 280 | 5/6 plate (250 grams) |
| 21 | change | 1 | fraction | -1 - -0.8 | Decrease it to zero |
| 22 | change | 2 | fraction | -0.9 - -0.5 | Decrease it to less than half |
| 23 | change | 3 | fraction | -0.6 - -0.1 | Decrease it a bit |
| 24 | change | 4 | fraction | 0 | No effect |
| 25 | change | 5 | fraction | 0.1 - 0.6 | Increase it a bit |
| 26 | change | 6 | fraction | 0.5 - 0.9 | Increase it over by half |
| 27 | change | 7 | fraction | 0.8 - 1.3 | Increase it over to double |
| 28 | change | 8 | fraction | -0.3 - 0.3 | Don't know |
Preprocessing
This code is used to preprocess the original questionnaire data from the above .csv file and to store the data as a usable variable to Opasnet base. The code stores a data.frame named survey.
Analyses
- Sketches about modelling determinants of eating (spring 2018) [4]
Figures, tables and stat analyses for the first manuscript
- Model run 8.5.2019 with thlVerse code (not run on Opasnet) [5] [6]
- Model run 26.8.2018 with fig 6 as table [7]
- Previous model runs are archived.
Methodological concerns:
- Is the warning in logistic regression important?
- Goodness of fit in logistic regression
- Calculate odds ratios in logistic regression
- You might treat independent ordinal variables as continuous
- Color blindness simulator to adjust colors for the color blind and for black and white printing
Luke data about fish consumption in Finland [8][9]
| Obs | Origin | Species | 1999 | 2000 | 2001 | 2002 | 2003 | 2004 | 2005 | 2006 | 2007 | 2008 | 2009 | 2010 | 2011 | 2012 | 2013 | 2014 | 2015 | 2016 | 2017 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | domestic fish | Total | 6.1 | 6.1 | 5.9 | 6.2 | 5.8 | 5.3 | 5.2 | 5.0 | 5.0 | 4.4 | 4.5 | 4.3 | 3.8 | 3.8 | 3.8 | 4.0 | 4.1 | 4.1 | 4.1 |
| 2 | domestic fish | Farmed rainbow trout | 1.6 | 1.6 | 1.6 | 1.6 | 1.3 | 1.3 | 1.4 | 1.1 | 1.1 | 1.2 | 1.2 | 1.1 | 1.0 | 1.0 | 1.1 | 1.1 | 1.3 | 1.2 | 1.2 |
| 3 | domestic fish | Baltic herring | 0.8 | 1.2 | 1.1 | 1.1 | 0.9 | 0.8 | 0.7 | 0.5 | 0.4 | 0.4 | 0.4 | 0.3 | 0.3 | 0.3 | 0.3 | 0.3 | 0.3 | 0.3 | 0.3 |
| 4 | domestic fish | Pike | 0.8 | 0.7 | 0.7 | 0.7 | 0.6 | 0.7 | 0.7 | 0.8 | 0.7 | 0.6 | 0.6 | 0.6 | 0.5 | 0.4 | 0.4 | 0.5 | 0.5 | 0.4 | 0.4 |
| 5 | domestic fish | Perch | 0.7 | 0.7 | 0.7 | 0.7 | 0.6 | 0.6 | 0.6 | 0.7 | 0.7 | 0.5 | 0.5 | 0.5 | 0.4 | 0.4 | 0.4 | 0.5 | 0.5 | 0.4 | 0.4 |
| 6 | domestic fish | Vendace | 0.7 | 0.7 | 0.7 | 0.7 | 0.8 | 0.8 | 0.7 | 0.6 | 0.6 | 0.6 | 0.6 | 0.7 | 0.6 | 0.6 | 0.6 | 0.6 | 0.6 | 0.5 | 0.6 |
| 7 | domestic fish | European whitefish | 0.4 | 0.4 | 0.4 | 0.4 | 0.3 | 0.3 | 0.3 | 0.3 | 0.5 | 0.3 | 0.3 | 0.3 | 0.3 | 0.3 | 0.3 | 0.3 | 0.2 | 0.3 | 0.3 |
| 8 | domestic fish | Pike perch | 0.3 | 0.2 | 0.2 | 0.2 | 0.3 | 0.3 | 0.3 | 0.3 | 0.3 | 0.3 | 0.3 | 0.3 | 0.3 | 0.3 | 0.3 | 0.3 | 0.3 | 0.4 | 0.4 |
| 9 | domestic fish | Other domestic fish | 0.8 | 0.6 | 0.5 | 0.8 | 1.0 | 0.5 | 0.5 | 0.7 | 0.7 | 0.5 | 0.6 | 0.5 | 0.4 | 0.5 | 0.4 | 0.5 | 0.4 | 0.5 | 0.5 |
| 10 | imported fish | Total | 6.0 | 6.2 | 7.0 | 7.1 | 8.0 | 8.6 | 7.9 | 8.6 | 9.7 | 9.7 | 9.3 | 10.2 | 11.1 | 10.9 | 10.8 | 10.9 | 10.2 | 9.1 | 9.8 |
| 11 | imported fish | Farmed rainbow trout | 0.2 | 0.3 | 0.4 | 0.6 | 0.9 | 0.6 | 0.6 | 0.7 | 1.0 | 1.0 | 0.8 | 0.8 | 0.9 | 1.0 | 0.9 | 0.9 | 0.8 | 0.9 | 0.8 |
| 12 | imported fish | Farmed salmon | 1.0 | 0.9 | 1.2 | 1.3 | 1.6 | 2.2 | 1.9 | 2.0 | 2.7 | 2.6 | 2.9 | 3.1 | 3.9 | 4.2 | 4.0 | 4.4 | 4.1 | 3.5 | 4.0 |
| 13 | imported fish | Tuna (prepared and preserved) | 1.0 | 1.2 | 1.4 | 1.4 | 1.5 | 1.6 | 1.6 | 1.5 | 1.7 | 1.7 | 1.6 | 1.7 | 1.7 | 1.6 | 1.9 | 1.7 | 1.6 | 1.4 | 1.5 |
| 14 | imported fish | Saithe (frozen fillet) | 0.7 | 0.6 | 0.7 | 0.7 | 0.7 | 0.6 | 0.4 | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 | 0.6 | 0.5 | 0.5 | 0.5 | 0.5 | 0.4 | 0.4 |
| 15 | imported fish | Shrimps | 0.5 | 0.4 | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 | 0.5 | 0.6 | 0.6 | 0.6 | 0.7 | 0.7 | 0.6 | 0.6 | 0.5 | 0.5 | 0.4 | 0.4 |
| 16 | imported fish | Herring and Baltic herring (preserved) | 0.6 | 0.5 | 0.6 | 0.6 | 0.5 | 0.4 | 0.5 | 0.6 | 0.5 | 0.6 | 0.3 | 0.5 | 0.4 | 0.3 | 0.4 | 0.5 | 0.5 | 0.5 | 0.5 |
| 17 | imported fish | Other imported fish | 2.0 | 2.3 | 2.2 | 2.0 | 2.3 | 2.7 | 2.4 | 2.8 | 2.7 | 2.7 | 2.6 | 2.9 | 2.9 | 2.7 | 2.5 | 2.3 | 2.3 | 1.9 | 2.2 |
Descriptive statistics
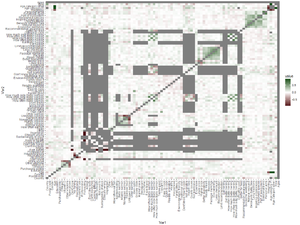
Model must contain predictors such as country, gender, age etc. Maybe we should first study what determinants are important? Model must also contain determinants that would increase or decrease fish consumption. This should be conditional on the current consumption. How? Maybe we should look at principal coordinates analysis with all questions to see how they behave.
Also look at correlation table to see clusters.
Some obvious results:
- If reports no fish eating, many subsequent answers are NA.
- No vitamins correlates negatively with vitamin intake.
- Unknown salmon correlates negatively with the types of salmon eaten.
- Different age categories correlate with each other.
However, there are also meaningful negative correlations:
- Country vs allergy
- Country vs Norwegian salmon and Rainbow trout
- Country vs not traditional.
- Country vs recommendation awareness
- Allergy vs economic wellbeing
- Baltic salmon use (4 questions) vs Don't like taste and Not used to
- All questions between Easy to cook ... Traditional dish
Meaningful positive correlations:
- All questions between Baltic salmon ... Rainbow trout
- How often Baltic salmon/herring/side salmon/side herring
- How much Baltic salmon/herring/side salmon/side herring
- Better availability ... Recommendation
- All questions between Economic wellbeing...Personal aims
- Omega3, Vitamin D, and Other vitamins
Model runs
- Model run 13.3.2017
- Model run 21.4.2017 [10] old code from Answer merged to this code and debugged
Bayes model
- Model run 3.3.2017. All variables assumed independent. [11]
- Model run 3.3.2017. p has more dimensions. [12]
- Model run 25.3.2017. Several model versions: strange binomial+multivarnormal, binomial, fractalised multivarnormal [13]
- Model run 27.3.2017 [14]
- Other models except multivariate normal were archived and removed from active code 29.3.2017.
- Model run 29.3.2017 with raw data graphs [15]
- Model run 29.3.2017 with salmon and herring ovariables stored [16]
- Model run 13.4.2017 with first version of coordinate matrix and principal coordinate analysis [17]
- Model run 20.4.2017 [18] code works but needs a safety check against outliers
- Model run 21.4.2017 [19] some model results plotted
- Model run 21.4.2017 [20] ovariables produced by the model stored.
- Model run 18.5.2017 [21] small updates
- 13.2.2018 old model run but with new Opasnet [22]
Initiate ovariables
jsp taken directly from data WITHOUT salmpling
Amount estimated from a bayesian model
- Model run 24.5.2017 [23]
Amount estimates directly from data rather than from a bayesian model
- Initiation run 18.5.2017 [24]
- Initiation run 24.2.2018: sampling from survey rather than each respondent once [25]
Initiate other ovariables
- Code stores ovariables assump, often, much, oftenside, muchside, amount.
- Model run 19.5.2017 [26]
- Initiation run 24.5.2017 without jsp [27]
- Model run 8.6.2017 [28]
Other code
This is code for analysing EFSA food intake data about fish for BONUS GOHERR manuscript Ronkainen L, Lehikoinen A, Haapasaari P, Tuomisto JT. 2019.
Dependencies
The survey data will be used as input in the benefit-risk assessment of Baltic herring and salmon intake, which is part of the WP5 work in Goherr-project.
See also
- Useful information about Wishart distribution and related topics:
Keywords
References
- ↑ 1.0 1.1 Mia Pihlajamäki, Arja Asikainen, Suvi Ignatius, Päivi Haapasaari and Jouni T. Tuomisto. Forage Fish as Food: Consumer Perceptions on Baltic Herring. Sustainability 2019, 11(16), 4298; https://doi.org/10.3390/su11164298
- ↑ Tuomisto, J.T., Asikainen, A., Meriläinen, P., Haapasaari, P. Health effects of nutrients and environmental pollutants in Baltic herring and salmon: a quantitative benefit-risk assessment. BMC Public Health 20, 64 (2020). https://doi.org/10.1186/s12889-019-8094-1
Related files