Opasnet base structure: Difference between revisions
(→Some useful syntax: indices updated, but not ready yet) |
(marked outdated) |
||
| (63 intermediate revisions by 4 users not shown) | |||
| Line 2: | Line 2: | ||
[[Category:Open assessment]] | [[Category:Open assessment]] | ||
[[Category:Tool]] | [[Category:Tool]] | ||
{{variable}} | {{variable|moderator = Jouni | ||
| reference = {{publication | |||
| authors = Juha Villman, Einari Happonen, Jouni T. Tuomisto | |||
| page = Opasnet Base structure | |||
| explanation = | |||
| publishingyear = 2010 | |||
| urn = | |||
| elsewhere = | |||
}} | |||
}} | |||
:''This page is about the '''old structure of Opasnet Base''' used in ca. 2008-2011. For a description about the current database, see [[Opasnet base 2]]. | |||
'''Opasnet | ==Question== | ||
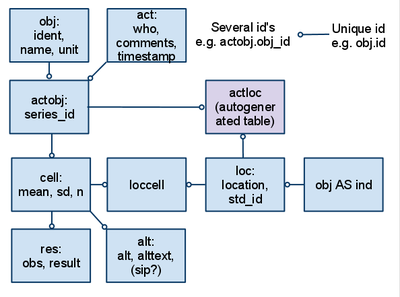
[[image:Opasnet Base structure.png|thumb|400px|Structure and connections (lines) of the tables (boxes) in the [[Opasnet Base]]. All table identifiers are called id (so they can be called by like obj.id). When obj.id is referred to in another table such as actobj, it is called actobj.obj_id. The latter end of a one-to-many relationship is marked with a ring. Important substantive fields are listed inside table boxes.]] | |||
'''Opasnet Base''' is a storage and retrieval system for [[result]]s of [[variable]] and [[data]] from [[study|studies]]. What is the structure of [[Opasnet Base]] such that it enables the following functionalities? | |||
# Storage of results of variables with uncertainties when necessary, and as multidimensional arrays when necessary.{{reslink|Should all variables go to result distribution database?}} | # Storage of results of variables with uncertainties when necessary, and as multidimensional arrays when necessary.{{reslink|Should all variables go to result distribution database?}} | ||
# Automatic retrieval of results when called from [[Opasnet wiki]] or other platforms or modelling systems. | # Automatic retrieval of results when called from [[Opasnet wiki]] or other platforms or modelling systems. | ||
# Description and handling of the [[ | # Description and handling of the [[index|indices]]s that a [[variable]] may take. | ||
# It is possible to protect some results and data from reading by unauthorised persons. | # It is possible to protect some results and data from reading by unauthorised persons. | ||
# If is possible to build user interfaces for easily entering observations into the Base. | # If is possible to build user interfaces for easily entering observations into the Base. | ||
==Answer== | |||
Opasnet base is a [[:en:MySQL|MySQL]] database located at http://base.opasnet.org. | |||
===Data=== | ===Data structure=== | ||
:''Main article: '''[[Data structures in Opasnet]]''''' | |||
All data should be convertible into the following format: | |||
==== | {| {{prettytable}} | ||
! colspan="3"| || colspan="3" style="background-color: #FFD8F0;"|Observation | |||
|----- | |||
! style="background-color: #CCDFC8;"|Year || style="background-color: #CCDFC8;"|Sex || style="background-color: #CCDFC8;"|Age || style="background-color: #DFB8D0;"|Height || style="background-color: #DFB8D0;"|Weight || style="background-color: #DFB8D0;"|Description | |||
|----- | |||
| style="background-color: #ECFFE8;"|2009 || style="background-color: #ECFFE8;"|Male || style="background-color: #ECFFE8;"|20 || style="background-color: #ECDFE8;"|178 || style="background-color: #ECDFE8;"|70 || style="background-color: #ECDFE8;"|An optional column for descriptive text about each row. | |||
|----- | |||
| style="background-color: #ECFFE8;"|2009 || style="background-color: #ECFFE8;"|Male || style="background-color: #ECFFE8;"|30 || style="background-color: #ECDFE8;"|174 || style="background-color: #ECDFE8;"|79 || style="background-color: #ECDFE8;"| | |||
|----- | |||
| style="background-color: #ECFFE8;"|2010 || style="background-color: #ECFFE8;"|Male || style="background-color: #ECFFE8;"|25 || style="background-color: #ECDFE8;"|183 || style="background-color: #ECDFE8;"|84 || style="background-color: #ECDFE8;"| | |||
|----- | |||
| style="background-color: #ECFFE8;"|2010 || style="background-color: #ECFFE8;"|Female || style="background-color: #ECFFE8;"|22 || style="background-color: #ECDFE8;"|168 || style="background-color: #ECDFE8;"|65 || style="background-color: #ECDFE8;"| | |||
|} | |||
where | |||
{| {{prettytable}} | |||
| style="background-color: #CCDFC8;"|Names of explanation columns, also known as indices. | |||
|----- | |||
| style="background-color: #ECFFE8;"|Explanation data, also known as locations. You can use these columns as search criteria. | |||
|----- | |||
| style="background-color: #FFD8F0;"|Observation index, typically called "Observation". Common name for all observation columns | |||
|----- | |||
| style="background-color: #DFB8D0;"|Names of observation columns. These are the parameters of interest. | |||
|----- | |||
| style="background-color: #ECDFE8;"|Observation data. These are the actual measurements. | |||
|----- | |||
|} | |||
===Table structure in the database=== | |||
==== All tables ==== | |||
=== | |||
{| VALIGN="top" BORDER="0" | {| VALIGN="top" BORDER="0" | ||
|- | |- | ||
| | |VALIGN="top"| | ||
{| | {| {{prettytable}} | ||
| | |colspan=5|'''act''' | ||
| | |- | ||
|colspan=5|'''Uploads, updates, and other actions''' | |||
|- | |||
|Field||Type||Null||Extra||Key | |||
|- | |||
|id||int(10) unsigned||NO||auto_increment||PRI | |||
|- | |||
|acttype_id||tinyint(3) unsigned||NO||||MUL | |||
|- | |||
|who||varchar(50)||NO|||| | |||
|- | |||
|comments||varchar(250)||YES|||| | |||
|- | |- | ||
| | |time||timestamp||NO|||| | ||
| | |||
| | |||
| | |||
| | |||
| | |||
| | |||
| | |||
|- | |- | ||
| | |temp_id||int(10) unsigned||NO||||MUL | ||
| | |||
| | |||
| | |||
|} | |} | ||
|VALIGN="top"| | |VALIGN="top"| | ||
{| | {| {{prettytable}} | ||
| | |colspan=5|'''actloc''' | ||
|- | |||
|colspan=5|'''Locations of an act''' | |||
|- | |||
|Field||Type||Null||Extra||Key | |||
|- | |- | ||
| | |actobj_id||int(10) unsigned||NO||||PRI | ||
|- | |- | ||
| | |loc_id||int(10) unsigned||NO||||PRI | ||
| | |||
| int(10) | |||
| | |||
| | |||
| | |||
| | |||
| | |||
| | |||
|} | |} | ||
|VALIGN="top"| | |VALIGN="top"| | ||
{| | {| {{prettytable}} | ||
| | |colspan=5|'''actobj''' | ||
|- | |||
|colspan=5|'''Acts of an object''' | |||
|- | |- | ||
| | |Field||Type||Null||Extra||Key | ||
|- | |||
|id||int(10) unsigned||NO||auto_increment||PRI | |||
|- | |||
|act_id||int(10) unsigned||NO||||MUL | |||
|- | |||
|obj_id||int(10) unsigned||NO||||MUL | |||
|- | |||
|series_id||int(10) unsigned||NO||||MUL | |||
|- | |||
|unit||varchar(64)||YES|||| | |||
|} | |||
|- | |- | ||
| | | | ||
| | {| {{prettytable}} | ||
|colspan=5|'''acttype''' | |||
| | |- | ||
| | |colspan=5|'''List of action types''' | ||
| | |||
| | |||
|- | |- | ||
| | |Field||Type||Null||Extra||Key | ||
| | |||
|- | |- | ||
| | |id||int(10) unsigned||NO||auto_increment||PRI | ||
|- | |- | ||
| | |acttype||varchar(250)||NO||||UNI | ||
| | |||
| | |||
| | |||
| | |||
| | |||
| | |||
| | |||
| | |||
|} | |} | ||
|VALIGN="top"| | |VALIGN="top"| | ||
{| | {| {{prettytable}} | ||
| | |colspan=5|'''cell''' | ||
|- | |- | ||
| | |colspan=5|'''Cells of an object''' | ||
|- | |- | ||
| | |Field||Type||Null||Extra||Key | ||
| | |- | ||
| | |id||int(12) unsigned||NO||auto_increment||PRI | ||
| | |- | ||
| id | |actobj_id||int(10) unsigned||NO||||MUL | ||
| int( | |- | ||
| | |mean||float||YES|||| | ||
| | |- | ||
| | |sd||float||NO|||| | ||
| int(10) | |- | ||
| | |n||int(10)||NO|||| | ||
|- | |- | ||
| | |sip||varchar(2000)||YES|||| | ||
| int(10) | |||
| | |||
| | |||
|} | |} | ||
|VALIGN="top"| | |VALIGN="top"| | ||
{| | {| {{prettytable}} | ||
| | |colspan=5|'''loc''' | ||
|- | |||
|colspan=5|'''Location information''' | |||
|- | |||
|Field||Type||Null||Extra||Key | |||
|- | |||
|id||int(10) unsigned||NO||auto_increment||PRI | |||
|- | |||
|std_id||int(10) unsigned||NO||||MUL | |||
|- | |||
|obj_id_i||int(10) unsigned||NO||||MUL | |||
|- | |- | ||
| | |location||varchar(100)||NO|||| | ||
|- | |- | ||
| | |roww||mediumint(8) unsigned||NO|||| | ||
|- | |||
|description||varchar(150)||NO|||| | |||
| | |||
| | |||
| | |||
| | |||
| | |||
| | |||
| | |||
| | |||
| | |||
| | |||
| | |||
| | |||
| | |||
| | |||
| | |||
| | |||
|} | |} | ||
|- | |||
| | |||
{| {{prettytable}} | |||
|colspan=5|'''loccell''' | |||
|- | |||
|colspan=5|'''Locations of a cell''' | |||
|- | |- | ||
| | |Field||Type||Null||Extra||Key | ||
| | |||
|- | |- | ||
| | |cell_id||int(10) unsigned||NO||||PRI | ||
|- | |- | ||
| | |loc_id||int(10) unsigned||NO||||PRI | ||
| | |||
| int(10) | |||
| | |||
| | |||
| | |||
| | |||
| | |||
| | |||
|} | |} | ||
|VALIGN="top"| | |VALIGN="top"| | ||
{| | {| {{prettytable}} | ||
| | |colspan=5|'''obj''' | ||
|- | |||
|colspan=5|'''Object information (all objects)''' | |||
|- | |||
|Field||Type||Null||Extra||Key | |||
|- | |||
|id||int(10) unsigned||NO||auto_increment||PRI | |||
|- | |- | ||
| | |ident||varchar(20)||NO||||UNI | ||
|- | |- | ||
| | |name||varchar(200)||NO|||| | ||
| | |- | ||
| | |objtype_id||tinyint(3) unsigned||NO||||MUL | ||
| | |- | ||
| | |page||int(10) unsigned||NO|||| | ||
| tinyint(3) | |- | ||
| | |wiki_id||tinyint(3) unsigned||NO|||| | ||
| | |||
| | |||
| | |||
| | |||
| | |||
|} | |} | ||
|VALIGN="top"| | |VALIGN="top"| | ||
{| | {| {{prettytable}} | ||
| | |colspan=5|'''objtype''' | ||
|- | |||
|colspan=5|'''Types of objects''' | |||
|- | |||
|Field||Type||Null||Extra||Key | |||
|- | |- | ||
| | |id||tinyint(3)||NO||||PRI | ||
|- | |- | ||
| | |objtype||varchar(30)||NO|||| | ||
| | |||
| | |||
| | |||
| | |||
| | |||
| | |||
| | |||
| | |||
|} | |} | ||
|- | |- | ||
| | |||
{| {{prettytable}} | |||
|colspan=5|'''res''' | |||
|- | |||
|colspan=5|'''Result distribution (actual values)''' | |||
|- | |||
|Field||Type||Null||Extra||Key | |||
|- | |||
|id||bigint(20) unsigned||NO||auto_increment||PRI | |||
|- | |||
|cell_id||int(12) unsigned||NO||||MUL | |||
|- | |||
|obs||int(10) unsigned||NO|||| | |||
|- | |||
|result||float||NO|||| | |||
|- | |||
|restext||varchar(250)||YES|||| | |||
|- | |||
|implausible||binary(1)||YES|||| | |||
|} | |||
|VALIGN="top"| | |VALIGN="top"| | ||
{| | {| {{prettytable}} | ||
| | |colspan=5|'''wiki''' | ||
|- | |||
|colspan=5|'''Wiki information''' | |||
|- | |||
|Field||Type||Null||Extra||Key | |||
|- | |||
|id||tinyint(3)||NO||||PRI | |||
|- | |- | ||
| | |url||varchar(255)||NO|||| | ||
|- | |- | ||
| | |wname||varchar(20)||NO|||| | ||
| | |} | ||
| | |} | ||
====Contents of selected tables==== | |||
{| | |||
| | |||
{| {{prettytable}} | |||
|+ Table objtype | |||
! id!! objtype | |||
|---- | |||
|| 1|| Variable | |||
|---- | |||
|| 2|| Study | |||
|---- | |||
|| 3|| Method | |||
|---- | |||
|| 4|| Assessment | |||
|---- | |---- | ||
| | || 5|| Class | ||
| | |||
| | |||
|---- | |---- | ||
| | || 6|| Index | ||
| | |||
| | |||
|---- | |---- | ||
| | || 7|| Nugget | ||
| | |||
| | |||
| | |||
|---- | |---- | ||
| | || 8|| Encyclopedia article | ||
| | |||
| | |||
|---- | |---- | ||
| | || 9|| Run | ||
| | |||
| | |||
|---- | |---- | ||
| | |} | ||
| | | | ||
{| {{prettytable}} | |||
|+ Table acttype | |||
! id|| acttype | |||
|---- | |---- | ||
| | || 1|| Start object | ||
| | |---- | ||
| | || 2|| Finish assessment | ||
| | |---- | ||
| | || 3|| Update formula | ||
| | |||
|- | |||
| | |||
| | |||
| | |||
|---- | |---- | ||
| | || 4|| Upload data (replace) | ||
| | |||
|---- | |---- | ||
| | || 5|| Upload data (append) | ||
| | |||
| | |||
|---- | |---- | ||
| | || 6|| Review scope | ||
| | |||
| | |||
|---- | |---- | ||
| | || 7|| Review definition | ||
| | |||
| | |||
|---- | |---- | ||
| | || 8|| Add object info | ||
| | |||
| | |||
|---- | |---- | ||
|} | |} | ||
|} | |} | ||
== | ==Rationale== | ||
===Data=== | |||
====Software==== | |||
Because Opasnet base will contain very large amounts of mostly numerical information, the state-of-the-art structure is a [[:en:SQL|SQL]] database. Because of its flexibility, ease of use, and cost, [[:en:MySQL|MySQL]] is an optimal choice among SQL software. In addition to the database software, a [[variable transfer protocol]] is needed on top of that so that the results of variables can be retrieved and new results stored either automatically by a calculating software, or manually by the user. Fancy presenting software can be built on top of the database, but that is not the topic of this page. | |||
====Storage and retrieval of results of variables==== | |||
The most important functionality is to store and retrieve the results of variables. Because variables may take very different forms (from a single value such as natural constant to an uncertain spatio-temporal concentration field over the whole Europe), the database must be very flexible. The basic solution is described in the [[variable]] page, and it is only briefly summarised here. The result is described as | |||
P(R|x<sub>1</sub>,x<sub>2</sub>,...) | |||
where P(R) is the probability distribution of the result and x<sub>1</sub> and x<sub>2</sub> are defining [[location]]s of an [[index]] where a particular P(R) applies. Typically locations are operationalised as discrete [[Index|indices]]. A variable must have at least one [[index]]. [[Uncertainty]] about the true value of the variable is operationalised as a random sample from the probability distribution, in such a way that the samples are located along an index ''Sample'', which is a list of integers 1,2,3...n, where n=number of samples. | |||
== | * [http://en.opasnet.org/en-opwiki/index.php?title=Opasnet_base&oldid=7856 Old description of the structure] | ||
===Dependencies=== | |||
* [[Opasnet structure]] | |||
* | * [[Open assessment]] | ||
* | |||
=== | ====Replacing some cells==== | ||
It is possible that there is a large data, where there is a need to update only a few cells while all others remain the same. How should this be done? There are a few potential alternatives. | |||
# Use the current replace functionality. Replace all cells but most of them with the original value. | |||
# Use a new act_type that is similar to the current append functionality. This should be understood in a way that if there are two (or more) identical cells (based on cell indices and locations), then the newest result is used and all older ones are discarded. (If the old ''append'' is used, then new info is just seen as a new row in the data table, not a replacement of an existing row. | |||
# Add a new field into the cell (?) table with an updated cell_id (in a similar way than act_id and series_id are used in the actobj table). This way, the new cell can automatically inherit all locations of the old cell. | |||
=== | ===Formula structure=== | ||
Now it has become clear that it is not enough to have samples of the result distributions. It must be possible to completely recalculate the result based on the information in the [[Opasnet Base]]. There are different approaches: | |||
* Calculate the result based on a formula that may refer to other variables called parents. This is a deterministic approach. | |||
* | * Calculate the result based on the marginal distribution and (conditional) rank correlations with parent variables. This is a probabilistic approach. | ||
* | |||
This approach requires new tables, namely Formula and Language. | |||
: {{comment|11|Do we need tables DIF and DIP like Uninet?|--[[User:Jouni|Jouni]] 21:50, 30 December 2009 (UTC)}} | |||
* DIP | |||
** DIP_node_id | |||
** DIP_parent_node_id | |||
** DIP_corr_coeff | |||
** DIP_parent_index | |||
* DIF | |||
** DIF_node_id | |||
** DIF_formula | |||
** DIF_varnames_in_formula | |||
===Universal Opasnet Base=== | |||
The idea of universal Opasnet Base says that it should be possible to store results in such a way that the results themselves are public but their interpretation is limited. For example, patient symptoms and clinical test results should be openly available for research, but information about whose results they are should be private. This can be achieved with the following database structure. | |||
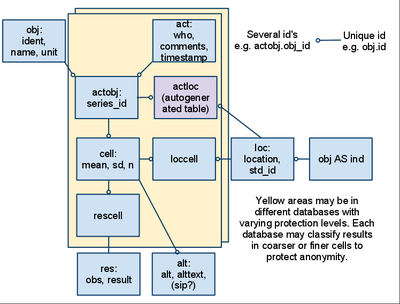
[[File:Universal Opasnet Base structure.png|thumb|400px|Universal Opasnet Base has some parts that exist in different versions depending on the privacy level. The yellow areas are e.g. a public area and a private area. The parts that are white are public.]] | |||
' | Let's say that it is enough to have two security levels, public and private. A person wants to record personal health information into the database. She logs in with her personal user name. The private profile gives the name (say, Liisa) and social security number of the person, while the public profile says only "30-40-year-old woman in Finland". Liisa writes down her symptoms or medical information and saves them. This is what is stored in the databases: | ||
{| {{prettytable}} | |||
|+ '''Information stored in the public and private databases. The private database can read tables from the public one but not vice versa. | |||
! Table, field | |||
! Private database | |||
! Public database | |||
|---- | |||
| act.who | |||
| Liisa, 010175-1024 | |||
| Woman, 30-40 a | |||
|---- | |||
| act.when | |||
| 2011-03-09 22:09:10 | |||
| 2011-03 | |||
|---- | |||
| obj.name | |||
| N/A. Data is taken from public side. | |||
| Pregnancy test | |||
|---- | |||
| loccell.loc_id (locations and indices explained) | |||
| Person = 010175-1024 <br>Time = 2011-03-09 <br>Test = Clearblue digital test | |||
| Age = 30-40 <br>Sex = Female <br>Country = Finland <br>Time = 2011-03 <br>Test = Clearblue digital test | |||
|---- | |||
| res.restext | |||
| N/A. Data is taken from public side. | |||
| Pregnant 1-2 weeks. | |||
|---- | |||
|} | |||
Based on the information, anyone can see that there is a woman in Finland who has used a Clearblue pregnancy test and the result was positive. But there is no way an outsider could connect this information to any particular person, because all information that could be used for linking is located in the private website. However, an authorised person from health case could see the data in the private database and connect Liisa and the test result. | |||
==See also== | |||
* [http://en.opasnet.org/en-opwiki/index.php?title=Opasnet_base_structure&oldid=18900#All_tables:_Overview A previous discussion about the structure] | |||
* [http://en.opasnet.org/en-opwiki/index.php?title=Opasnet_base_structure&oldid=18900#Main_tables A previous structure and related discussions] | |||
* [http://en.opasnet.org/en-opwiki/index.php?title=Opasnet_base_structure&oldid=18900#Tasks_performed Previous tasks performed] | |||
; A basic query for retrieving the full result of a variable upload (an example): | |||
{{#sql-query: | |||
SELECT obj.ident, obj.name, obj.unit, obj.page, obj.wiki_id, comments, mean, sd, n, location, ind.ident, obs, result, restext | |||
FROM obj | |||
LEFT JOIN actobj ON actobj.obj_id = obj.id | |||
LEFT JOIN act ON actobj.act_id = act.id | |||
LEFT JOIN cell ON cell.actobj_id = actobj.id | |||
LEFT JOIN loccell ON loccell.cell_id = cell.id | |||
LEFT JOIN loc on loccell.loc_id = loc.id | |||
LEFT JOIN obj AS ind ON loc.obj_id_i = ind.id | |||
LEFT JOIN res ON res.cell_id = cell.id | |||
WHERE obj.ident = "Op_en1912" | |||
AND actobj.series_id = 190 | |||
LIMIT 0,100 | |||
}} | |||
;Some useful syntax | |||
* http://www.baycongroup.com/sql_join.htm | |||
* [[:image:Opasnet base connection.ANA|Opasnet base connection.ANA]] for Analytica: for writing and reading variable results into and from the database. Writing requires a password. For SQL used in the model, see the model page. | |||
* [http://en.opasnet.org/en-opwiki/index.php?title=Opasnet_base&oldid=7181#Other_queries Some historical queries] | |||
* [http://en.opasnet.org/en-opwiki/index.php?title=Opasnet_base_structure&oldid=14214#Some_useful_syntax Some historical queries 2] | |||
[[Category:Opasnet Base]] | |||
[[Category:Data]] | |||
[[Category:Opasnet]] | |||
Latest revision as of 12:01, 10 January 2014
| [show] |
|---|
- This page is about the old structure of Opasnet Base used in ca. 2008-2011. For a description about the current database, see Opasnet base 2.
Question
Opasnet Base is a storage and retrieval system for results of variable and data from studies. What is the structure of Opasnet Base such that it enables the following functionalities?
- Storage of results of variables with uncertainties when necessary, and as multidimensional arrays when necessary.R↻
- Automatic retrieval of results when called from Opasnet wiki or other platforms or modelling systems.
- Description and handling of the indicess that a variable may take.
- It is possible to protect some results and data from reading by unauthorised persons.
- If is possible to build user interfaces for easily entering observations into the Base.
Answer
Opasnet base is a MySQL database located at http://base.opasnet.org.
Data structure
- Main article: Data structures in Opasnet
All data should be convertible into the following format:
| Observation | |||||
|---|---|---|---|---|---|
| Year | Sex | Age | Height | Weight | Description |
| 2009 | Male | 20 | 178 | 70 | An optional column for descriptive text about each row. |
| 2009 | Male | 30 | 174 | 79 | |
| 2010 | Male | 25 | 183 | 84 | |
| 2010 | Female | 22 | 168 | 65 | |
where
| Names of explanation columns, also known as indices. |
| Explanation data, also known as locations. You can use these columns as search criteria. |
| Observation index, typically called "Observation". Common name for all observation columns |
| Names of observation columns. These are the parameters of interest. |
| Observation data. These are the actual measurements. |
Table structure in the database
All tables
|
|
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
|
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Contents of selected tables
|
|
Rationale
Data
Software
Because Opasnet base will contain very large amounts of mostly numerical information, the state-of-the-art structure is a SQL database. Because of its flexibility, ease of use, and cost, MySQL is an optimal choice among SQL software. In addition to the database software, a variable transfer protocol is needed on top of that so that the results of variables can be retrieved and new results stored either automatically by a calculating software, or manually by the user. Fancy presenting software can be built on top of the database, but that is not the topic of this page.
Storage and retrieval of results of variables
The most important functionality is to store and retrieve the results of variables. Because variables may take very different forms (from a single value such as natural constant to an uncertain spatio-temporal concentration field over the whole Europe), the database must be very flexible. The basic solution is described in the variable page, and it is only briefly summarised here. The result is described as
P(R|x1,x2,...)
where P(R) is the probability distribution of the result and x1 and x2 are defining locations of an index where a particular P(R) applies. Typically locations are operationalised as discrete indices. A variable must have at least one index. Uncertainty about the true value of the variable is operationalised as a random sample from the probability distribution, in such a way that the samples are located along an index Sample, which is a list of integers 1,2,3...n, where n=number of samples.
Dependencies
Replacing some cells
It is possible that there is a large data, where there is a need to update only a few cells while all others remain the same. How should this be done? There are a few potential alternatives.
- Use the current replace functionality. Replace all cells but most of them with the original value.
- Use a new act_type that is similar to the current append functionality. This should be understood in a way that if there are two (or more) identical cells (based on cell indices and locations), then the newest result is used and all older ones are discarded. (If the old append is used, then new info is just seen as a new row in the data table, not a replacement of an existing row.
- Add a new field into the cell (?) table with an updated cell_id (in a similar way than act_id and series_id are used in the actobj table). This way, the new cell can automatically inherit all locations of the old cell.
Formula structure
Now it has become clear that it is not enough to have samples of the result distributions. It must be possible to completely recalculate the result based on the information in the Opasnet Base. There are different approaches:
- Calculate the result based on a formula that may refer to other variables called parents. This is a deterministic approach.
- Calculate the result based on the marginal distribution and (conditional) rank correlations with parent variables. This is a probabilistic approach.
This approach requires new tables, namely Formula and Language.
- ----11: . Do we need tables DIF and DIP like Uninet? --Jouni 21:50, 30 December 2009 (UTC) (type: truth; paradigms: science: comment)
- DIP
- DIP_node_id
- DIP_parent_node_id
- DIP_corr_coeff
- DIP_parent_index
- DIF
- DIF_node_id
- DIF_formula
- DIF_varnames_in_formula
Universal Opasnet Base
The idea of universal Opasnet Base says that it should be possible to store results in such a way that the results themselves are public but their interpretation is limited. For example, patient symptoms and clinical test results should be openly available for research, but information about whose results they are should be private. This can be achieved with the following database structure.
Let's say that it is enough to have two security levels, public and private. A person wants to record personal health information into the database. She logs in with her personal user name. The private profile gives the name (say, Liisa) and social security number of the person, while the public profile says only "30-40-year-old woman in Finland". Liisa writes down her symptoms or medical information and saves them. This is what is stored in the databases:
| Table, field | Private database | Public database |
|---|---|---|
| act.who | Liisa, 010175-1024 | Woman, 30-40 a |
| act.when | 2011-03-09 22:09:10 | 2011-03 |
| obj.name | N/A. Data is taken from public side. | Pregnancy test |
| loccell.loc_id (locations and indices explained) | Person = 010175-1024 Time = 2011-03-09 Test = Clearblue digital test |
Age = 30-40 Sex = Female Country = Finland Time = 2011-03 Test = Clearblue digital test |
| res.restext | N/A. Data is taken from public side. | Pregnant 1-2 weeks. |
Based on the information, anyone can see that there is a woman in Finland who has used a Clearblue pregnancy test and the result was positive. But there is no way an outsider could connect this information to any particular person, because all information that could be used for linking is located in the private website. However, an authorised person from health case could see the data in the private database and connect Liisa and the test result.
See also
- A previous discussion about the structure
- A previous structure and related discussions
- Previous tasks performed
- A basic query for retrieving the full result of a variable upload (an example)
{{#sql-query: SELECT obj.ident, obj.name, obj.unit, obj.page, obj.wiki_id, comments, mean, sd, n, location, ind.ident, obs, result, restext FROM obj LEFT JOIN actobj ON actobj.obj_id = obj.id LEFT JOIN act ON actobj.act_id = act.id LEFT JOIN cell ON cell.actobj_id = actobj.id LEFT JOIN loccell ON loccell.cell_id = cell.id LEFT JOIN loc on loccell.loc_id = loc.id LEFT JOIN obj AS ind ON loc.obj_id_i = ind.id LEFT JOIN res ON res.cell_id = cell.id WHERE obj.ident = "Op_en1912" AND actobj.series_id = 190 LIMIT 0,100 }}
- Some useful syntax
- http://www.baycongroup.com/sql_join.htm
- Opasnet base connection.ANA for Analytica: for writing and reading variable results into and from the database. Writing requires a password. For SQL used in the model, see the model page.
- Some historical queries
- Some historical queries 2